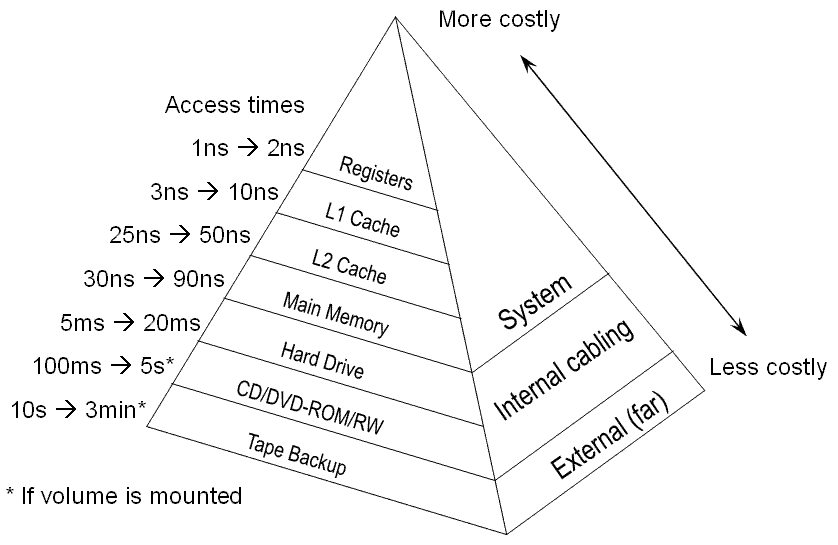
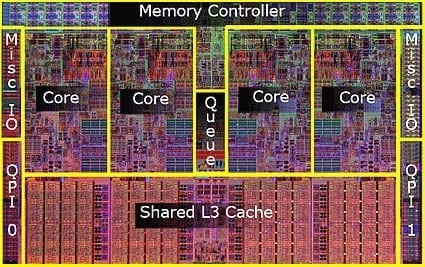
- Big ideas: Memory, CPUs, and Networking
- Let's draw some pictures!
https://tinyurl.com/rsmas-python2
- Somewhere between 1 and 16 cores
- Modern processors will support hyperthreading / virtual cores
Source and Jeff Dean, Peter Norvig, etc.
| Event | Nanoseconds | Microseconds | Milliseconds | Comparison |
|---|---|---|---|---|
| L1 cache reference | 0.5 | - | - | - |
| L2 cache reference | 7.0 | - | - | 14x L1 cache |
| Main memory reference | 100.0 | - | - | 20x L2 cache, 200x L1 cache |
| Compress 1K bytes with Zippy | 3,000.0 | 3 | - | - |
| Send 1K bytes over 1 Gbps network | 10,000.0 | 10 | - | - |
| Read 4K randomly from SSD | 150,000.0 | 150 | - | ~1GB/sec SSD |
| Read 1 MB sequentially from memory | 250,000.0 | 250 | - | - |
| Round trip within same datacenter | 500,000.0 | 500 | - | - |
| Read 1 MB sequentially from SSD | 1,000,000.0 | 1,000 | 1 | ~1GB/sec SSD, 4X memory |
| Disk seek | 10,000,000.0 | 10,000 | 10 | 20x datacenter roundtrip |
| Read 1 MB sequentially from disk | 20,000,000.0 | 20,000 | 20 | 80x memory, 20X SSD |
| Send packet CA → Netherlands → CA | 150,000,000.0 | 150,000 | 150 | - |
1 ns = 10^-9 seconds 1 us = 10^-6 seconds = 1,000 ns 1 ms = 10^-3 seconds = 1,000 us = 1,000,000 ns
Lets multiply all these durations by a billion:
L1 cache reference 0.5 s One heart beat (0.5 s)
L2 cache reference 7 s Long yawn
Main memory reference 100 s Brushing your teeth
Compress 1K bytes with Zippy 50 min One episode of a TV show (including ad breaks)
Send 2K bytes over 1 Gbps network 5.5 hr From lunch to end of work day
SSD random read 1.7 days A normal weekend
Read 1 MB sequentially from memory 2.9 days A long weekend
Round trip within same datacenter 5.8 days A medium vacation
Read 1 MB sequentially from SSD 11.6 days Waiting for almost 2 weeks for a delivery
Disk seek 16.5 weeks A semester in university
Read 1 MB sequentially from disk 7.8 months Almost producing a new human being
The above 2 together 1 year
Send packet CA->Netherlands->CA 4.8 years Average time it takes to complete a bachelor's degree
- CPUs are really fast
- The goal: Have the CPU always be working
- Memory is fast-ish, hard drives are slow
- You want to make your CPU happy by always giving it data that is close at hand
- Networks are reeeeaaally slow
- Since there's so much downtime when we need to get data from disk or over a network, we can use the CPU for another task
- Switching between tasks can keep our CPU working
- 3,000-30,000 ns for a context switch, or about 50 minutes to 8 hours in human time
- To allow this, we want multiple different programs that are able to run at the same time
- These are called processes
- A process has 4 parts
- The program code
- The Call Stack (what is actually happening in the program)
- Variables, files, memory, etc
- Within a program, we have also have multiple mini-programs running
- These are called threads
- A child thread and its parent share the same memory (usually)
- Running a script from the command line
import subprocess
subprocess.call(['python3 run_me.py', shell=True])- Getting the stdout and stderr from a process
output, errors = subprocess.Popen(['ls', '-la', '*.py'],
stdout=subprocess.PIPE).communicate()- Piping processes
p1 = subprocess.Popen(["cat", "file.log"], stdout=subprocess.PIPE)
p2 = subprocess.Popen(["tail", "-1"], stdin=p1.stdout, stdout=subprocess.PIPE)
p1.stdout.close() # Allow p1 to receive a SIGPIPE if p2 exits.
output,err = p2.communicate()- Lots of models
- Hard to do super efficiently
- However, there are some easy ways to get gains
- Using many cores from your machines is different than using many cores on multiple machines
- Pegasus, as far as I can tell, makes it as if you are using a lot of cores on one machine
- Vectorizing functions is the easiest step to wriitng fast and parallel code
- Many Numpy Operations already are parallelized across machines
- Split
- Think about how to break up the data so that you can compute the same thing on smaller parts of the larger dataset
- Apply
- Solve the problem a bunch of times on smaller versions of your large data
- Combine
- Then combine those mini solutions into one big solution
- In image analysis, you can apply convolutions because they only matter on a small subset of the data
- If you need all of the data to do each computation, it's less efficient
# Parallel version is equivalent to the following
def map_parallel(func, input_data):
intermediate_data = []
for i in input_data:
intermediate_data.append(func(i)
return intermediate_data
def reduce(func, intermediate_data):
starting_value = None
for value in intermediate_data:
if starting_value == None:
starting_value = value
else:
starting_value = func(starting_value, value)
return value- Prepare data to be split between processors
- Use
multiprocessingto applymapstep - Combine the results of the
mapresult as needed - Easy when your data is small
- Good for simple functions
- Some good examples
from multiprocessing import Pool
def f(x):
return x*x
if __name__ == '__main__':
with Pool(5) as p:
print(p.map(f, [1, 2, 3]))
###################################
# Sequential
results = []
for item in my_array:
results.append(my_function(item))
# Parallel
from multiprocessing.dummy import Pool as ThreadPool
pool = ThreadPool(4)
results = pool.map(my_function, my_array)- For more complicated parallel tasks
- Converts functions to C code
- Can parallelize code easily (if you read the documentation :D )
@numba.jit(nopython=True, parallel=True)
def logistic_regression(Y, X, w, iterations):
for i in range(iterations):
w -= np.dot(((1.0 / (1.0 + np.exp(-Y * np.dot(X, w))) - 1.0) * Y), X)
return w- Virtual environments: https://conda.io/docs/user-guide/tasks/manage-environments.html
- Figuring out your dependencies:
pip freeze pylint <filename>for lintingautopep8 <filename>for correcting easy linting errors
- Two types
- Unit Test
- Just tests individual functions
- Define some input and some desired output
- Tests whether it works
- Integration Test
- Combining functions
- COmbining modules
- Very simple example cases
- Unit Test
- Tests are written in a separate folder usually