You signed in with another tab or window. Reload to refresh your session.You signed out in another tab or window. Reload to refresh your session.You switched accounts on another tab or window. Reload to refresh your session.Dismiss alert
we want to balance workload to make threads as busy as possible
we may (or may not have) GPUs available
we are using xgboost hist (CPU or GPU) with a known hyperparameter combo (with RNG seed + a bit of sub sampling / column sampling)
Provides the following, by run:
total time to train
model training time
time to build training matrix to feed to xgboost
time to build testing matrix to feed to xgboost
prediction time
garbage collection time (R specific overhead)
metric
Also in addition, auto clustering is performed to find if there are potential changes in the data series.
Expected behavior:
Script success (no OOM)
No increasing memory usage once stabilized (gc must work as it is)
Lot of outliers during early/end of parallelization (in timings) due to firing up/off threads simultaneously
Stable metric overall
Scaling up with CPU threads and GPUs (throughput metric: more models per second done)
Scaling up parallel should be better than scaling up multithreaded
Get some nice output CSV/chart files for analysis
Dominating Model Training time, followed by Model Predict time (ignoring Metric time)
Objective of the libraries tested:
Go through the matrix creation (for the model) to the model training (train a machine learning model) to the prediction (predict quickly) as fast as possible, while using the least memory possible (we are not using fork, to make sure NUMA is affecting the least possible the parallel processes)
Keep metric performance as high as possible against a blind spot (no early stopping) with usually good but fine out of the box hyperparameters (GBDT: 500 rounds, 0.05 learning rate, 6 depth, 0.9 row sampling, 0.9 column sampling by tree) which actually should favor speed for all libraries tested
Example charts with 1 to 4 GPUs:
1 GPU (20.436s / model):
2 GPUs (10.666s / model):
3 GPUs (6.999s / model):
4 GPUs (5.182s / model) :
I am currently using a shell script to run a big batch of different works on my Dual Xeon 6154 with 4 Quadro P1000 (4 Quadro P1000 is approximately equal to 1 GeForce 1080). It gets the current folder, and executes the R benchmark script with different parameters.
For the 0.1m dataset used and if you want to use GPU, you need about 1GB GPU RAM peak per process. Do not believe it just use the peak 659 MB reported by nvidia-smi, the peak is very short and happens at the beginning of training (running 4 models is the max for 4GB RAM GPU, not well optimized implementation for sparse data in xgboost GPU hist).
Run
Parallel Threads
Model Threads
Parallel GPUs
GPU Threads
Models
1
1
1
1
1
25
2
2
1
2
1
50
3
3
1
3
1
100
4
4
1
4
1
250
5
4
1
1
4
50
6
8
1
2
4
100
7
12
1
3
4
250
8
16
1
4
4
500
9
1
1
0
0
25
10
9
1
0
0
50
11
18
1
0
0
100
12
35
1
0
0
250
13
70
1
0
0
500
14
1
1
0
0
50
15
1
9
0
0
50
16
1
18
0
0
50
17
1
35
0
0
50
18
1
70
0
0
50
(yeah, run 9 and 14 are identical...)
Shell script, you will need in the folder you are running the benchmark:
Note my script clears up GOMP_CPU_AFFINITY variable if you set it. If you set it and you are not clearing it, you will end up with only 1 thread being used for all runs (and all processes will share the same thread).
The full bench_xgb_test.R script. Use bench_xgb_test -h to get help for the commands. Passing --args=true as parameter allows you to check the parameters you passed to the script, without executing any training / data loading. You may run want it interactively if you wish...
Required R packages:
optparse (parses the CLI parameters)
data.table (load data, xgboost data holders)
parallel (parallel sockets for multiprocessing on available threads)
xgboost (with GPU support if you want to use GPU parameters)
Matrix (xgboost specific)
ggplot2 (for charting)
ClusterR (for the chart, autoclustering time series by time aka model number)
Script:
# Sets OpenMP to 1 thread by default, bypasses xgboost forcing all thread on xgb.DMatrix
Sys.setenv(OMP_NUM_THREADS=1)
suppressMessages({
library(optparse)
library(data.table)
library(parallel)
library(xgboost)
library(Matrix)
})
args_list<-list(
optparse::make_option("--parallel_threads", type="integer", default=1, metavar="Parallel CPU Threads",
help="Number of threads for parallel training for CPU (automatically changed if using GPU), should be greater than or equal to parallel_gpus * gpus_threads [default: %default]"),
optparse::make_option("--model_threads", type="integer", default=1, metavar="Model CPU Threads",
help="Number of threads for training a single model, total number of threads is parallel_threads * model_threads [default: %default]"),
optparse::make_option("--parallel_gpus", type="integer", default=0, metavar="Parallel GPU Threads",
help="Number of GPUs to use for parallel training, use 0 for no GPU [default: %default]"),
optparse::make_option("--gpus_threads", type="integer", default=0, metavar="Model GPU Threads",
help="Number of parallel models to train per GPU (uses linearly more RAM), use 0 for no GPU [default: %default]"),
optparse::make_option("--number_of_models", type="integer", default=1, metavar="Number of Models",
help="Number of models to train in total [default: %default]"),
optparse::make_option("--wkdir", type="character", default="", metavar="Working Directory",
help="The working directory, do NOT forget it! [default: \"%default\"]"),
optparse::make_option("--train_file", type="character", default="", metavar="Training File",
help="The training file to use relative to the working directory (or an absolute path), do NOT forget it! [default: \"%default\"]"),
optparse::make_option("--test_file", type="character", default="", metavar="Testing file",
help="The testing file to use relative to the working directory (or an absolute path), do NOT forget it! [default: \"%default\"]"),
optparse::make_option("--output_dir", type="character", default="", metavar="Output Directory",
help="The output directory for files (or an absolute path), do NOT forget it! [default: \"%default\"]"),
optparse::make_option("--output_csv", type="logical", default=TRUE, metavar="Output CSV File",
help="Outputs results as a CSV file [default: %default]"),
optparse::make_option("--output_chart", type="character", default="jpeg", metavar="Plot File Format",
help="Outputs results as a chart using the desired format, can be any of: \"none\" (for no chart), \"eps\", \"ps\", \"tex\" (pictex), \"pdf\", \"jpeg\", \"tiff\", \"png\", \"bmp\", \"svg\", \"wmf\" (Windows only) [default: \"%default\"]"),
optparse::make_option("--args", type="logical", default=FALSE, metavar="Argument Check",
help="Prints the arguments passed to the R script and exits immediately [default: %default]")
)
# Force data.table as 1 thread in case you are using Fork instead of Sockets (gcc: fork X in process Y when process Y used OpenMP once, fork X cannot use OpenMP otherwise it hangs forever)data.table::setDTthreads(1)
if (interactive()) {
# Put some parameters if you wish to test once...my_gpus<-1Lmy_gpus_threads<-1Lmy_threads<-parallel::detectCores() -1Lmy_threads_in_threads<-1Lmy_runs<-100Lmy_train<-"train-0.1m.csv"my_test<-"test.csv"my_output<-"./output"my_csv<-TRUEmy_chart<-"jpeg"# my_cpu <- system("lscpu | sed -nr '/Model name/ s/.*:\\s*(.*) @ .*/\\1/p' | sed ':a;s/ / /;ta'")# CHANGE: 0.1M = GPU about 958 MB at peak... choose wisely (here, we are putting 4 models per GPU)if (my_gpus>0L) {
# my_threads <- min(my_gpus * my_gpus_threads, my_threads)my_threads<-my_gpus*my_gpus_threads
}
} else {
# Old school method... obsolete# DIR="$( cd "$( dirname "${BASH_SOURCE[0]}" )" && pwd )"# Rscript bench_file.R 1 1 0 0 25 ${DIR} ../train-0.1m.csv ../test.csv# args <- commandArgs(trailingOnly = TRUE)## setwd(args[6])# my_gpus <- args[3]# my_gpus_threads <- args[4]# my_threads <- args[1]# my_threads_in_threads <- args[2]# my_runs <- args[5]# my_train <- args[7]# my_test <- args[8]# DIR="$( cd "$( dirname "${BASH_SOURCE[0]}" )" && pwd )"# Rscript bench_xgb_test.R --parallel_threads=1 --model_threads=1 --parallel_gpus=0 --gpus_threads=0 --number_of_models=25 --wkdir=${DIR} --train_file=../train-0.1m.csv --test_file=../test.csv --output_dir=./output --output_csv=TRUE --output_chart=jpeg --args=TRUE# Rscript bench_xgb_test.R --parallel_threads=1 --model_threads=1 --parallel_gpus=0 --gpus_threads=0 --number_of_models=25 --wkdir=${DIR} --train_file=../train-0.1m.csv --test_file=../test.csv --output_dir=./output --output_csv=TRUE --output_chart=jpegargs<-optparse::parse_args(optparse::OptionParser(option_list=args_list))
setwd(args$wkdir)
my_gpus<-args$parallel_gpusmy_gpus_threads<-args$gpus_threadsmy_threads<-args$parallel_threadsmy_threads_in_threads<-args$model_threadsmy_runs<-args$number_of_modelsmy_train<-args$train_filemy_test<-args$test_filemy_output<-args$output_dirmy_csv<-args$output_csvmy_chart<-args$output_chartif (my_gpus>0L) {
# my_threads <- min(my_gpus * my_gpus_threads, my_threads)my_threads<-my_gpus*my_gpus_threadsargs$parallel_threads<-my_threads
}
if (args$args) {
print(args)
stop("\rArgument check done.")
}
}
# Load data and do preprocessing
cat("[", format(Sys.time(), "%a %b %d %Y %X"), "] [Data] Loading data.\n", sep="")
d_train<- fread(my_train, showProgress=FALSE)
d_test<- fread(my_test, showProgress=FALSE)
invisible(gc(verbose=FALSE))
cat("[", format(Sys.time(), "%a %b %d %Y %X"), "] [Data] Transforming data.\n", sep="")
X_train_test<- sparse.model.matrix(dep_delayed_15min~.-1, data= rbindlist(list(d_train, d_test))); invisible(gc(verbose=FALSE))
n1<- nrow(d_train)
n2<- nrow(d_test)
X_train<-X_train_test[1L:n1,]; invisible(gc(verbose=FALSE))
X_test<-X_train_test[(n1+1L):(n1+n2),]; invisible(gc(verbose=FALSE))
labels_train<- as.numeric(d_train$dep_delayed_15min=="Y")
labels_test<- as.numeric(d_test$dep_delayed_15min=="Y")
# dxgb_train <- xgb.DMatrix(data = X_train, label = labels_train); invisible(gc(verbose = FALSE))# dxgb_test <- xgb.DMatrix(data = X_test); invisible(gc(verbose = FALSE))
rm(d_train, d_test, X_train_test, n1, n2); invisible(gc(verbose=FALSE))
# CHANGE: metric functionmetric<-function(preds, labels) {
x1<- as.numeric(preds[labels==1])
n1<- as.numeric(length(x1))
x2<- as.numeric(preds[labels==0])
n2<- as.numeric(length(x2))
r<- rank(c(x1,x2))
return((sum(r[1:n1]) -n1* (n1+1) /2) / (n1*n2))
}
# CHANGE: trainer functiontrainer<-function(x, row_sampling, col_sampling, max_depth, n_iter, learning_rate, nbins, nthread, n_gpus, gpu_choice, objective) {
matrix_train_time<- system.time({
dxgb_train<-xgboost::xgb.DMatrix(data=X_train, label=labels_train)
})[[3]]
matrix_test_time<- system.time({
dxgb_test<-xgboost::xgb.DMatrix(data=X_test, label=labels_test)
})[[3]]
if (n_gpus>0) {
model_time<- system.time({
set.seed(x)
model_train<-xgboost::xgb.train(data=dxgb_train,
objective=objective,
nrounds=n_iter,
max_depth=max_depth,
eta=learning_rate,
subsample=row_sampling,
colsample_bytree=col_sampling,
nthread=nthread,
n_gpus=n_gpus,
gpu_id=gpu_choice,
tree_method="gpu_hist",
max_bin=nbins,
predictor="gpu_predictor")
})[[3]]
} else {
model_time<- system.time({
set.seed(x)
model_train<-xgboost::xgb.train(data=dxgb_train,
objective=objective,
nrounds=n_iter,
max_depth=max_depth,
eta=learning_rate,
subsample=row_sampling,
colsample_bytree=col_sampling,
nthread=nthread,
n_gpus=0,
tree_method="hist",
max_bin=nbins)
})[[3]]
}
pred_time<- system.time({
model_predictions<- predict(model_train, newdata=dxgb_test)
})[[3]]
perf<- metric(preds=model_predictions, labels=labels_test)
rm(model_train, model_predictions, dxgb_train, dxgb_test)
gc_time<- system.time({
invisible(gc(verbose=FALSE))
})[[3]]
return(list(matrix_train_time=matrix_train_time, matrix_test_time=matrix_test_time, model_time=model_time, pred_time=pred_time, gc_time=gc_time, perf=perf))
}
# Parallel Section
cat("[", format(Sys.time(), "%a %b %d %Y %X"), "]", " [Parallel] ", my_threads, " Process(es) Creation Time: ", sprintf("%04.03f", system.time({cl<- makeCluster(my_threads)})[[3]]), "s\n", sep="")
cat("[", format(Sys.time(), "%a %b %d %Y %X"), "]", " [Parallel] Sending Hardware Specifications Time: ", sprintf("%04.03f", system.time({clusterExport(cl=cl, c("my_threads", "my_gpus", "my_threads_in_threads"))})[[3]]), "s\n", sep="")
invisible(parallel::parLapply(cl=cl, X= seq_len(my_threads), function(x) {
Sys.sleep(time=my_threads/20) # Prevent file clash on many core systems (typically 50+ threads might attempt to read exactly at the same time the same file, especially if the disk is slow)
suppressPackageStartupMessages(library(xgboost))
suppressPackageStartupMessages(library(Matrix))
suppressPackageStartupMessages(library(data.table))
id<<-x
}))
cat("[", format(Sys.time(), "%a %b %d %Y %X"), "]", " [Parallel] Sending Data Time: ", sprintf("%04.03f", system.time({clusterExport(cl=cl, c("trainer", "metric", "X_train", "X_test", "labels_train", "labels_test", "my_threads"))})[[3]]), "s\n", sep="")
# Having issues? In a CLI: sudo pkill Rtime_finish<- system.time({
time_all<-parallel::parLapplyLB(cl=cl, X= seq_len(my_runs), function(x) {
if (my_gpus==0L) {
gpus_to_use<-0gpus_allowed<-0
} else {
gpus_to_use<- (id-1) %%my_gpusgpus_allowed<-1
}
speed_out<- system.time({
speed_in<- trainer(x=x,
row_sampling=0.9,
col_sampling=0.9,
max_depth=6,
n_iter=500,
learning_rate=0.05,
nbins=255,
nthread=my_threads_in_threads,
n_gpus=gpus_allowed,
gpu_choice=gpus_to_use,
objective="binary:logistic")
})[[3]]
rm(gpus_to_use)
return(list(total=speed_out, matrix_train_time=speed_in$matrix_train_time, matrix_test_time=speed_in$matrix_test_time, model_time=speed_in$model_time, pred_time=speed_in$pred_time, gc_time=speed_in$gc_time, perf=speed_in$perf))
})
})[[3]]
# Clearup all R sessions from this process, except the master
stopCluster(cl)
closeAllConnections()
rm(cl, metric, trainer, X_train, X_test, labels_train, labels_test); invisible(gc(verbose=FALSE))
cat("[", format(Sys.time(), "%a %b %d %Y %X"), "]", " [Parallel] Total Time: ", sprintf("%04.03f", time_finish), "s\n", sep="")
# Gather Data# Get datatime_total<- unlist(lapply(time_all, function(x) {round(x[[1]], digits=3)}))
matrix_train_time<- unlist(lapply(time_all, function(x) {round(x[[2]], digits=3)}))
matrix_test_time<- unlist(lapply(time_all, function(x) {round(x[[3]], digits=3)}))
model_time<- unlist(lapply(time_all, function(x) {round(x[[4]], digits=3)}))
pred_time<- unlist(lapply(time_all, function(x) {round(x[[5]], digits=3)}))
gc_time<- unlist(lapply(time_all, function(x) {round(x[[6]], digits=3)}))
perf<- unlist(lapply(time_all, function(x) {round(x[[7]], digits=6)}))
# Put all data togethertime_table<- data.table(Run= seq_len(my_runs),
time_total=time_total,
matrix_train_time=matrix_train_time,
matrix_test_time=matrix_test_time,
model_time=model_time,
pred_time=pred_time,
gc_time=gc_time,
perf=perf)
if (my_csv) {
fwrite(time_table, paste0(my_output, "/ml-perf_xgb_gbdt_", substr(my_train, 1, nchar(my_train) -4), "_", my_threads, "Tx", my_threads_in_threads, "T_", my_gpus, "GPU_", my_runs, "m_", sprintf("%04.03f", time_finish), "s.csv"))
}
# Analyze Dataif (my_chart!="none") {
suppressMessages({
library(ggplot2)
library(ClusterR)
})
# Create time series matrixtime_table_matrix<- apply(as.matrix(time_table[, 2:8, with=FALSE]), MARGIN=2, function(x) {
y<- cumsum(x)
y/ max(y)
})
# Compute optimal number of non-parametric clustersclusters<- Optimal_Clusters_Medoids(data=time_table_matrix,
max_clusters=2:10,
distance_metric="manhattan",
criterion="silhouette",
threads=1,
swap_phase=TRUE,
verbose=FALSE,
plot_clusters=FALSE,
seed=1)
# Compute clustersclusters_selected<- Cluster_Medoids(data=time_table_matrix,
clusters=1+ which.max(unlist(lapply(clusters, function(x) {x[[3]]}))),
distance_metric="manhattan",
threads=1,
swap_phase=TRUE,
verbose=FALSE,
seed=1)
time_table[, Cluster:= as.character(clusters_selected$clusters)]
# Melt datatime_table_vertical<- melt(time_table, id.vars= c("Run", "Cluster"), measure.vars= c("time_total", "matrix_train_time", "matrix_test_time", "model_time", "pred_time", "gc_time", "perf"), variable.name="Variable", value.name="Value", variable.factor=FALSE, value.factor=FALSE)
# Rename melted variables to have details in charttime_table_vertical[Variable=="time_total", Variable:= paste0("1. Total Time (Σ=", sprintf("%04.03f", sum(Value)), ", μ=", sprintf("%04.03f", mean(Value)), ", σ=", sprintf("%04.03f", sd(Value)), ")")]
time_table_vertical[Variable=="model_time", Variable:= paste0("2. Model Time (Σ=", sprintf("%04.03f", sum(Value)), ", μ=", sprintf("%04.03f", mean(Value)), ", σ=", sprintf("%04.03f", sd(Value)), ")")]
time_table_vertical[Variable=="matrix_train_time", Variable:= paste0("3. Matrix Train Build Time (Σ=", sprintf("%04.03f", sum(Value)), ", μ=", sprintf("%04.03f", mean(Value)), ", σ=", sprintf("%04.03f", sd(Value)), ")")]
time_table_vertical[Variable=="matrix_test_time", Variable:= paste0("4. Matrix Test Build Time (Σ=", sprintf("%04.03f", sum(Value)), ", μ=", sprintf("%04.03f", mean(Value)), ", σ=", sprintf("%04.03f", sd(Value)), ")")]
time_table_vertical[Variable=="pred_time", Variable:= paste0("5. Predict Time (Σ=", sprintf("%04.03f", sum(Value)), ", μ=", sprintf("%04.03f", mean(Value)), ", σ=", sprintf("%04.03f", sd(Value)), ")")]
time_table_vertical[Variable=="gc_time", Variable:= paste0("6. Garbage Collector Time (Σ=", sprintf("%04.03f", sum(Value)), ", μ=", sprintf("%04.03f", mean(Value)), ", σ=", sprintf("%04.03f", sd(Value)), ")")]
time_table_vertical[Variable=="perf", Variable:= paste0("7. Metric (Σ=", sprintf("%07.06f", sum(Value)), ", μ=", sprintf("%07.06f", mean(Value)), ", σ=", sprintf("%07.06f", sd(Value)), ")")]
cat(sort(unique(time_table_vertical$Variable)), sep="\n")
# Plot a nice chartmy_plot<- ggplot(data=time_table_vertical, aes(x=Run, y=Value, group=Cluster, color=Cluster)) + geom_point() + facet_wrap(facets=Variable~., nrow=4, ncol=2, scales="free_y") + labs(title="'Performance' over Models", subtitle= paste0(my_runs, " Models over ", sprintf("%04.03f", time_finish), " seconds using ", my_threads, " threads and ", my_gpus, " GPUs (Throughput: ", sprintf("%04.03f", time_finish/my_runs), "s / Model", ")"), x="Model", y="Value or Time (s)") + theme_bw() + theme(legend.position="none")
ggsave(filename= paste0(my_output, "/ml-perf_xgb_gbdt_", substr(my_train, 1, nchar(my_train) -4), "_", my_threads, "Tx", my_threads_in_threads, "T_", my_gpus, "GPU_", my_runs, "m_", sprintf("%04.03f", time_finish), "s.jpg"),
plot=my_plot,
device=my_chart,
width=24,
height=16,
units="cm",
dpi="print")
if (interactive()) {
print(my_plot)
}
}
cat("[", format(Sys.time(), "%a %b %d %Y %X"), "] Done computations. Quitting R.\n", sep="")
The text was updated successfully, but these errors were encountered:
Script example for benchmark where we have:
Provides the following, by run:
Also in addition, auto clustering is performed to find if there are potential changes in the data series.
Expected behavior:
Objective of the libraries tested:
Example charts with 1 to 4 GPUs:
1 GPU (20.436s / model):
2 GPUs (10.666s / model):
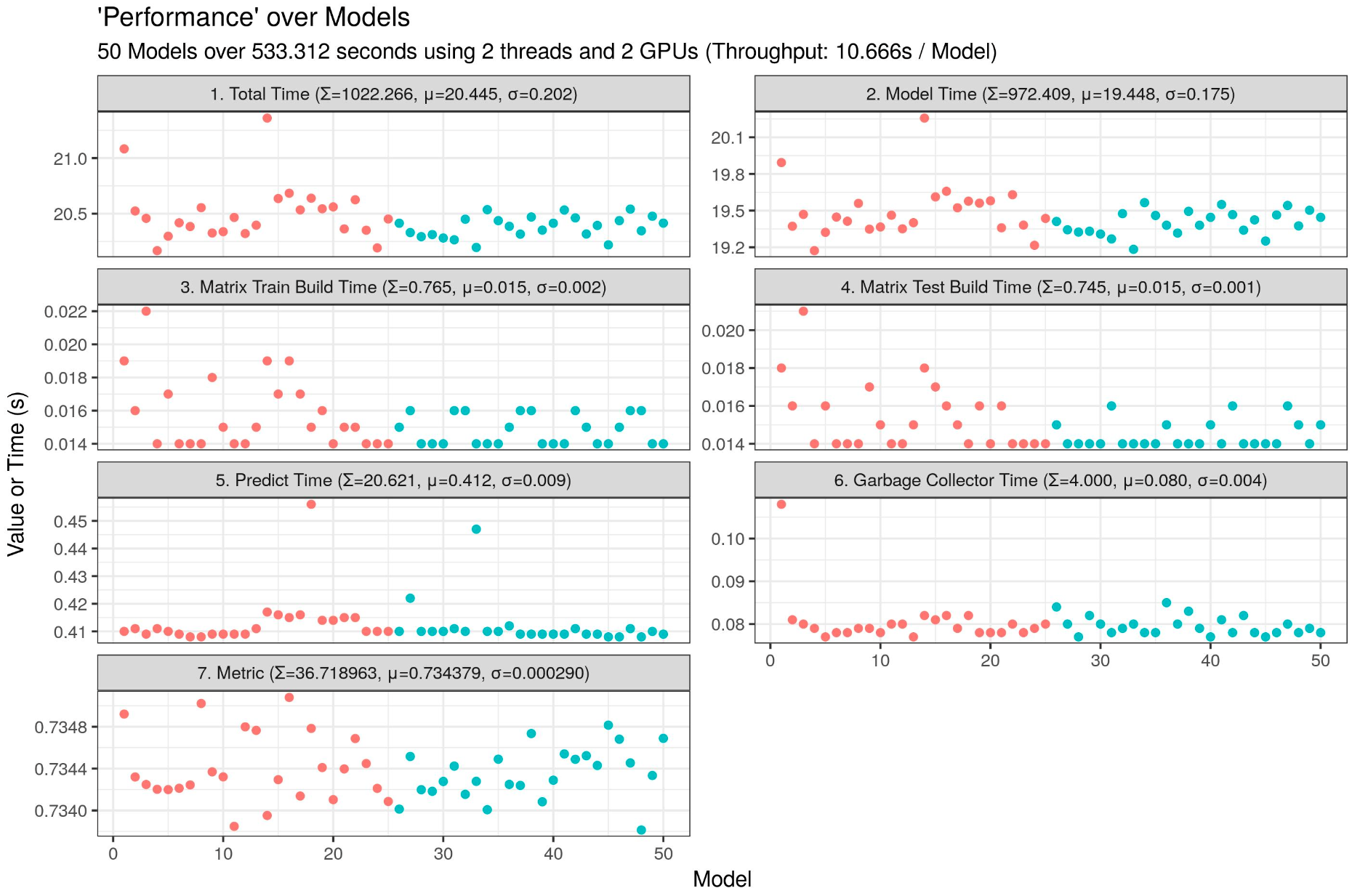
3 GPUs (6.999s / model):
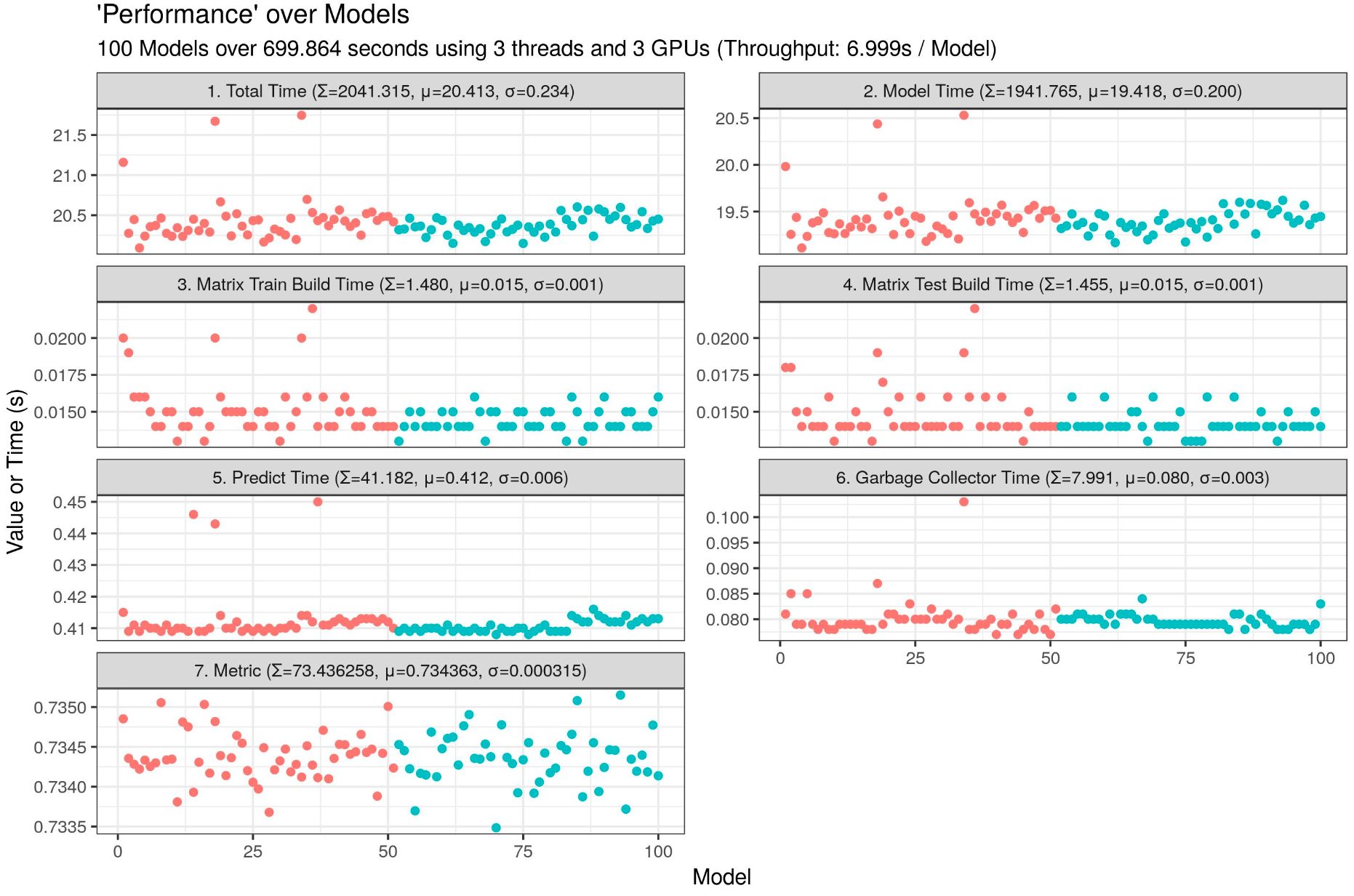
4 GPUs (5.182s / model) :
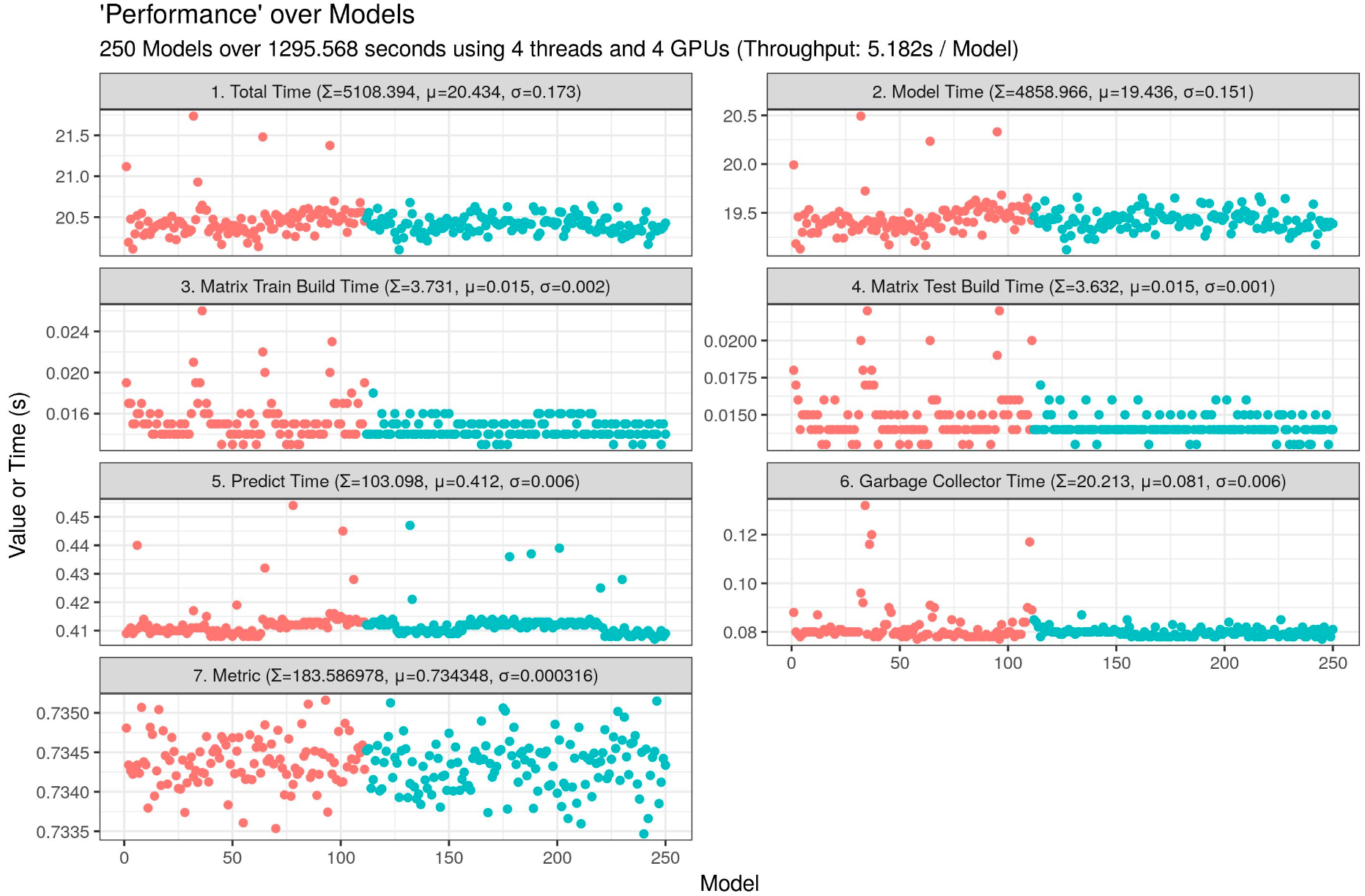
I am currently using a shell script to run a big batch of different works on my Dual Xeon 6154 with 4 Quadro P1000 (4 Quadro P1000 is approximately equal to 1 GeForce 1080). It gets the current folder, and executes the R benchmark script with different parameters.
For the 0.1m dataset used and if you want to use GPU, you need about 1GB GPU RAM peak per process. Do not believe it just use the peak 659 MB reported by
nvidia-smi, the peak is very short and happens at the beginning of training (running 4 models is the max for 4GB RAM GPU, not well optimized implementation for sparse data in xgboost GPU hist).(yeah, run 9 and 14 are identical...)
Shell script, you will need in the folder you are running the benchmark:
Note my script clears up GOMP_CPU_AFFINITY variable if you set it. If you set it and you are not clearing it, you will end up with only 1 thread being used for all runs (and all processes will share the same thread).
You should see similar to this during training:
The full bench_xgb_test.R script. Use
bench_xgb_test -hto get help for the commands. Passing--args=trueas parameter allows you to check the parameters you passed to the script, without executing any training / data loading. You may run want it interactively if you wish...Required R packages:
Script:
The text was updated successfully, but these errors were encountered: