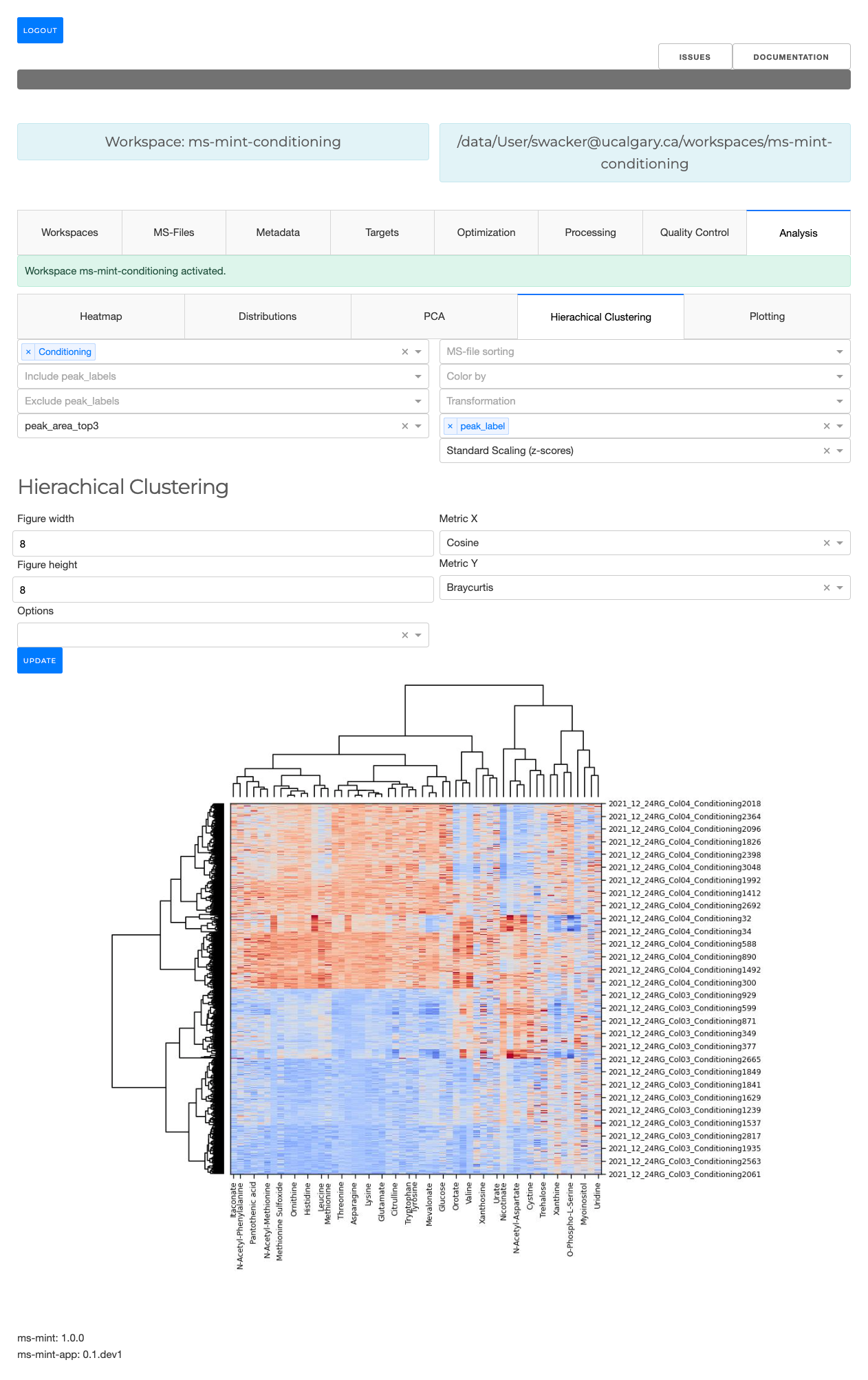
The Metabolomics Integrator (MINT) is a post-processing tool for liquid chromatography-mass spectrometry (LCMS) based metabolomics. Metabolomics is the study of all metabolites (small chemical compounds) in a biological sample e.g. from bacteria or a human blood sample. The metabolites can be used to define biomarkers used in medicine to find treatments for diseases or for the development of diagnostic tests or for the identification of pathogens such as methicillin resistant Staphylococcus aureus (MRSA). More information on how to install and run the program can be found in the Documentation or check out the Tutorial to jump right into it.
Starting with version 1.0.0, we have updated the installation setup to use pyproject.toml. Additionally, the main script to start Mint has been changed from Mint.py to Mint. Furthermore, each release of the repository will now be assigned a DOI to facilitate citation of the software.
-
Brown K, Thomson CA, Wacker S, Drikic M, Groves R, Fan V, et al. Microbiota alters the metabolome in an age- and sex- dependent manner in mice. Nat Commun. 2023;14: 1348.
-
Ponce LF, Bishop SL, Wacker S, Groves RA, Lewis IA. SCALiR: A Web Application for Automating Absolute Quantification of Mass Spectrometry-Based Metabolomics Data. Anal Chem. 2024;96: 6566–6574.
You can find installation instructions here
All contributions, bug reports, code reviews, bug fixes, documentation improvements, enhancements, and ideas are welcome. This includes recommendations for software architecture, code design, and efficiency improvements.
The project follows PEP8 standard and uses Black and Flake8 to ensure a consistent code format throughout the project.
To get in touch, please open a GitHub issue.
This project would not be possible without the help of the open-source community. The tools and resources provided by GitHub, Docker-Hub, the Python Package Index, as well the answers from dedicated users on Stackoverflow and the Plotly community, as well as the free open-source packages used are the foundation of this project. Several people have made direct contributions to the codebase and we are extremely grateful for that.
- @rokm refactored the specfile for
Pyinstallerto create a windows package. - @bucknerns helped with the configuration of the
versioneerfile.
Last but not least, we want to thank all the users and early adopters that drive the development with feature requests and bug reports.