-
Notifications
You must be signed in to change notification settings - Fork 12
How It Works
- Flow Chart
- Main Functions and Files Description
- Initial Data Preparation
- User Data Settings
- Species Data Settings
- DetEdit Interface
- Data Analysis
The following diagram shows the processing pipeline to get all the parameters and run the interface:
| Function Name | Description |
|---|---|
| detEdit | Launches the interface and returns user decisions in files grouping the detection labels in different annotation MAT files (TD – true detections, FD – false detections, ID – detection types |
| myDataSettings | Users’ specific parameters to run detEdit, mkLTSAsessions, modDet, and summaryParams. Parameters can be specified to overwrite default parameters and species default parameters. |
| Edetect, Edetect_wav | Simple energy threshold detector to detect signals from audio files in wave file format (.wav; Edetect_wav) or extended wave file format (.x.wav; Edetect). |
| mkLTSAsessions | Makes LTSAs per bout from LTSA of the corresponding deployment. Returns vector containing start times of spectral averages and power spectral densities into *LTSA.mat files. |
| make_TPWS | Groups detections in bouts and returns click parameters into *TPWS.mat files. |
| modDet | Deletes false detections stored in *FD.mat files from *TPWS.mat files and generates new *TPWS2.mat files. Optionally deletes ID detections stored in *ID.mat files as well. |
| initSpParams | Sets up visualization parameters for specific species. Parameters for eleven species of odontocetes are predefined. |
| summaryParams | Plots peak-to-peak amplitude, peak frequency, inter-click-interval histograms, and daily and weekly presence. |
| File Name | Description |
|---|---|
TPWS.mat |
Parameters of detected signals. Vector of detections start times (MTT), vector of peak-to-peak received sound pressure levels (MPP), matrix of time series (MSN), and matrix of power spectra (MSP). |
LTSA.mat |
LTSA parameters per bout. Matrix of binned power spectral densities (pwr) and vector of start times associated with each bin (pt). |
.ltsa |
File compression for audio data based on averaging spectra over long-term periods |
TD.mat |
Vector of true detection times |
FD.mat |
Vector of false detection times |
ID.mat |
Matrix of ID detection classified based on color ID color code. 1st column contains detection times and 2nd column the ID label |
Create parameter files required to use the interface:
LTSA files (.ltsa) are generated from a collection of WAV or XWAV files by averaging spectra over long-time periods and arranging these spectra sequentially as frequency-time spectrogram plots. LTSA files provide a quick overview of long-term recordings.
Using the Command Window in MATLAB, the .ltsafile is created as follows:
> mkLtsaand follow the prompt windows to specify the audio file format, file directories, and parameters (e.g., time average length and frequency bin size to average the data).
Only 5 filename formats of WAV and XWAV files are supported:
-
yymmdd-HHMMSS: SiteName_190326-123700.wav -
yymmdd_HHMMSS: SiteName_190326_123700.wav -
yyyymmdd_HHMMSS: SiteName_20190326_123700.wav -
yymmddHHMMSS: SiteName_190326123700.wav -
yyyymmddTHHMMS: SiteName_20190326T123700.wav
A *_TPWS.mat file contains the following matrices of detected signal parameters:
| Variable | Description |
|---|---|
MTT |
Vector of start times of detections |
MPP |
Vector of received level amplitudes (dBpp) |
MSP |
Matrix of detection spectra, where columns are dictated by the size of the fast-Fourier transform (FFT) used to generate the spectra each row represents a unique detection. |
MSN |
Matrix of waveforms, where columns are dictated by the window length used to extract the time series and each row represents a unique detection. |
Create *_TPWS.mat file is required to call the interface, so the user can:
-
provide start times of detected acoustic signals to create the
*_TPWS.matfile as follows:> edit make_TPWSIn the editor window, modify input/output locations and detection parameters to run the script.
-
apply a generic detector if you do not have any acoustic detections:
-
for use with XWAV files:
> EdetectThe user is prompted to supply inputs including a text file or spreadsheet containing detection parameters (see GOM_BW_pfile_320.xlsx for example), and a directory containing XWAV files.
-
for use with WAV files:
> Edetect_wav ('paramFile','E:\DetEdit\GOM_BW_pfile_320.xlsx',... 'tfFile','E:\MyTransferFunctionFiles\example_sig1_invSensit.tf',... 'timeFile','E:\DetEdit\BW_Effort.xlsx',... 'wavDir','E:\MyWAVFiles',... 'channel',1)
-
After *_TPWS.mat files have been created, the following step is making the snippets of LTSAs corresponding to bouts of detections saved in TPWS files.
A *_LTSA.mat file contains the following matrices of detected signal parameters:
| Variable | Description |
|---|---|
pt |
Vector of start times of spectral averages |
pwr |
Matrix of power spectral density averages, where columns are dictated by the step size used to average frequency spectrum bins. |
Using a script containing the user data settings (see myDataSettings for example), the LTSA snippets for each detection bout is performed as follows:
> mkLTSAsessions(@myDataSettings)or you can invoke the function directly
> mkLTSAsessionsand it will prompt you with a window to select the data settings script.
Assumming mkLTSAsessions invocation, the data settings scripts should contain the required following variables:
Before invoking any of the analysis (mkLTSAsessions, detEdit, modDet, or summaryParams), create your data settings script to define directories and parameters. Examples of data settings scripts are found in DetEdit\Settings folder. You can make different versions of these templates, with different names for different species, sites or analysis.
| Variable | Description | mkLTSAsessions | detEdit | modDet | summaryParams |
|---|---|---|---|---|---|
filePrefix |
TPWS.mat files name to match |
✔️ | ✔️ | ✔️ | ✔️ |
iterationNum |
Iteration number | ✔️ | ✔️ | ✔️ | ✔️ |
sampleRate |
Your data sample rate | ✔️ | ✔️ | ✔️ | ✔️ |
sp |
Species code | ✔️ | ✔️ | ✔️ | ✔️ |
tpwsDir |
TPWS.mat files directory |
✔️ | ✔️ | ✔️ | ✔️ |
tfName |
Transfer function file (.tf) directory |
🔶 | 🔶 | 🔶 | 🔶 |
ltsaDir |
LTSA.mat files directory |
✔️ |
✔️ = required variable 🔶 = optional variable (only if transfer function was not previously applied)
More parameter preferences can be specified to override default parameters (see myDataSettings for more details). Uncomment the parameters as needed to overwrite the default settings specified in initDefaultParams.
DetEdit is provided with initSpParams with signal-specific parameters for the 4 different analysis (mkLTSAsessions, detEdit, modDet, and summaryParams). Different odontocete species parameters and other signals are given in initSpParams
Species abbreviation code:
-
-
De: Delphinid -
Pm: Physeter macrorhynchus - Sperm whale -
Zc: Ziphius cavirostris - Cuvier’s beaked whale -
Me: Mesoplodon europaeus - Gervais' beaked whale -
BWG: BWG signal type - Unidentified beaked whale -
Md: BW31 signal type - Unidentified beaked whale -
Ko: Kogia spp. -
Po: Porpoise spp. -
Dl: Delphinapterus leucas - Beluga whale -
Mm: Monodon monoceros - Narwhal -
MFA: Mid-frequency active sonar
If given a species code, the specific parameter preferences will override the default parameters (see parameters in initDefaultParams for more details). If you define different parameter values in myDataSettings, those will overwrite the species-specific parameters (initSpParams) and the default parameters (initDefaultParams).
Using a script containing your data settings (see myDataSettings for example), you can invoke the interface as follows:
> detEdit(@myDataSettings)or you can invoke the interface directly
> detEditand it will prompt you with a window to select the data settings script.
While running, the user will be presented with progress information and at the beginning will be asked to specify Starting session, use 1 to start with the first bout.
After, the interface panels will be displayed. Arrange the panel as you wish for better visualization:
Annotation files ( *_TD.mat, *_FD.mat, and *_ID.mat) are created if executing detEdit for the first time for a *_TPWS.mat file.
All detections (stored in TPWS.mat file) are displayed in the interface. The annotations or classification you make would be stored in the corresponding annotation files, which will be updated each time a bout is edited.
| Annotation File | Description | Color detections |
|---|---|---|
*_TD.mat |
Vector of true detection times | |
*_FD.mat |
Vector of false detection times | |
*_ID.mat |
Matrix of ID detection classified based on color ID color code. 1st column contains detection times and 2nd column the ID label |
All annotation files from this stage are stored in the same directory where *_TPWS.mat files are located.
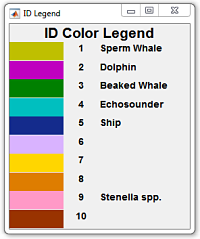
ID detection color legend:
| ID Label | RGB Color |
|---|---|
| 1 |  |
| 2 |  |
| 3 |  |
| 4 |  |
| 5 |  |
| 6 |  |
| 7 |  |
| 8 |  |
| 9 |  |
| 10 |  |
A species label can be given to each ID value label. Default is given at initDefaultParams:
You can modify the default species labels in myDataSettings in the following line:
line 77: paramsUser.mySpID = {'Label1','Label2','Label3', 'Label4', 'Label5', 'Label6', 'Label7', 'Label8', 'Label9', 'Label10'};The annotation process can be done per bout by annotating the entire window at once or by selecting batches of detections using the Matlab's paintbrush tool - (yellow default color).
click at the plot you want to brush the data and then press p. Brush button will appear selected and you will be ready to flag detections. You can select a different color for the brush which has different meanings.
Keyboard shortcuts:
| Keyboard | Brush color | Description |
|---|---|---|
t |
All detections in current window as true | |
f |
All detections in current window as false | |
u |
Update window contents | |
space |
Jump to next consecutive bout (next window session) | |
b |
Jump back one bout (previous window session) | |
r
|
u
|
Detections selected as false |
g
|
u
|
Detections selected as true |
y
|
Highlights and displays summary info in black outlined ( |
|
1) |
 + + u
|
Detections selected as ID color code |
Other keyboard shortcuts to modify the interface settings, this will prompt the user to introduce the desired number:
| Keyboard | Description |
|---|---|
j |
Jump to a non-consecutive bout |
a |
Adjust LTSA contrast and brightness |
s |
Adjust maximum time between detection scale |
h |
Adjust peak frequency scale |
d |
Adjust received levels (dBpp ) scale |
m |
Adjust transformed received levels (dBrms ) scale |
| '<' | Change transformed received levels (dBrms ) threshold |
: |
Change received levels (dBpp ) threshold |
^ |
Change peak frequency threshold |
To estimate the dataset's false positive rate, the following keyboard shortcuts will set the interface to display random detections for evaluation:
-
x: evaluation at the signal level, where random individual detections are displayed. -
w: evaluation at the time-bin interval level, where a batch of detections within the defined interval length is displayed and the user has to evaluate if at least one detection within the interval is true.
Then, the evaluation decision is done by pressing the following keyboard shortcuts:
-
x: as true detection -
z: as false detection
The random detections are selected systematically based on a given step size. The user can specify this parameter at myDataSettings, by uncommenting the line as follows:
line 42: paramsUser.c4fd = your number;this will overwrite the default step size parameter (see initDefaultParams for more details).
All evaluation decisions are stored in the *_TD.mat files.
After manually editing the detections using the interface, the user can remove the annotated detections as false positive detections stored in FD.mat file from the *_TPWS.mat files as follows:
> modDet(@myDataSettings)or you can invoke the function directly
> modDetand it will prompt you with a window to select the data settings script.
This will create TPWS2.mat files stored in a new folder (TPWS2 folder) within the current folder. The ID.mat files will be saved in the TPWS2 folder to maintain the ID detection labels.
Optionally, ID detections stored in ID.mat files can be excluded as follows:
line 71: paramsUser.excludeID = 1; % yes - 1 | no - 0. Exclude ID times from MTT filesor parsed out into sets using custom scripts.
Three different acoustic parameters, peak-to-peak received sound pressure levels, time-interval between detections, and peak frequencies from the detections of the modified file can be computed if desired as follows:
line paramsUser.calcParams = 1;% yes - 1 | no - 0. Calculate Parameters peak-to-peak RLs, inter-detection-interval and peak frequency The process of using detEdit and modDet can be repeated iteratively until all detections are annotated, or a sufficiently low percentage of false detections is obtained. For every iteration, new detection and annotation files are generated and stored together in a common directory to keep track of all changes.
A data summary of all the data from one site or from all the files with the same filePrefix can be computed as follows:
> summaryParams(@myDataSettings)or you can invoke the function directly
> summaryParamsand it will prompt you with a window to select the data settings script.
Histogram plots of peak-to-peak received sound pressure levels, time-interval between detections, and a time series of the weekly presence of the true detections at the signal level and the time-bin interval level will be displayed and stored in the corresponding directory.
You would be required to specify an excel file (see Test dataset as an example) with the monitoring effort start and end times, and the reference time you want the time series plot to start. You can specify these parameters in myDataSettings in the following lines:
line 27: effortTimes = 'E:\Effort.xls'; % specify excel file with effort times
line 28: referenceTime = '2010-04-01'; %reference time format 'yyyy-MM-dd'This software is Copyright © 2019 The Regents of the University of California. All Rights Reserved.