diff --git a/README.md b/README.md
index f8977715c..607bf8d95 100644
--- a/README.md
+++ b/README.md
@@ -65,6 +65,7 @@ PaddleScience 是一个基于深度学习框架 PaddlePaddle 开发的科学计
| 受力分析 | [3D 连接件变形](https://paddlescience-docs.readthedocs.io/zh/latest/zh/examples/bracket) | 机理驱动 | MLP | 无监督学习 | [Data](https://paddle-org.bj.bcebos.com/paddlescience/datasets/bracket/bracket_dataset.tar) | [Tutorial](https://docs.nvidia.com/deeplearning/modulus/modulus-v2209/user_guide/foundational/linear_elasticity.html) |
| 受力分析 | [结构震动模拟](https://paddlescience-docs.readthedocs.io/zh/latest/zh/examples/phylstm) | 机理驱动 | PhyLSTM | 监督学习 | [Data](https://paddle-org.bj.bcebos.com/paddlescience/datasets/PhyLSTM/data_boucwen.mat) | [Paper](https://arxiv.org/abs/2002.10253) |
| 受力分析 | [2D 弹塑性结构](https://paddlescience-docs.readthedocs.io/zh/examples/epnn.md) | 机理驱动 | EPNN | 无监督学习 | [Train Data](https://paddle-org.bj.bcebos.com/paddlescience/datasets/epnn/dstate-16-plas.dat)
[Eval Data](https://paddle-org.bj.bcebos.com/paddlescience/datasets/epnn/dstress-16-plas.dat) | [Paper](https://arxiv.org/abs/2204.12088) |
+| 受力分析和逆问题 | [3D 汽车控制臂变形](https://paddlescience-docs.readthedocs.io/zh/latest/zh/examples/control_arm.md) | 机理驱动 | MLP | 无监督学习 | - | - |
| 拓扑优化 | [2D 拓扑优化](https://paddlescience-docs.readthedocs.io/zh/latest/zh/examples/topopt.md) | 数据驱动 | TopOptNN | 监督学习 | [Data](https://paddle-org.bj.bcebos.com/paddlescience/datasets/topopt/top_dataset.h5) | [Paper](https://arxiv.org/pdf/1709.09578) |
diff --git a/docs/index.md b/docs/index.md
index 42139479c..469e08635 100644
--- a/docs/index.md
+++ b/docs/index.md
@@ -60,6 +60,7 @@
| 受力分析 | [3D 连接件变形](./zh/examples/bracket.md) | 机理驱动 | MLP | 无监督学习 | [Data](https://paddle-org.bj.bcebos.com/paddlescience/datasets/bracket/bracket_dataset.tar) | [Tutorial](https://docs.nvidia.com/deeplearning/modulus/modulus-v2209/user_guide/foundational/linear_elasticity.html) |
| 受力分析 | [结构震动模拟](./zh/examples/phylstm.md) | 机理驱动 | PhyLSTM | 监督学习 | [Data](https://paddle-org.bj.bcebos.com/paddlescience/datasets/PhyLSTM/data_boucwen.mat) | [Paper](https://arxiv.org/abs/2002.10253) |
| 受力分析 | [2D 弹塑性结构](./zh/examples/epnn.md) | 机理驱动 | EPNN | 无监督学习 | [Train Data](https://paddle-org.bj.bcebos.com/paddlescience/datasets/epnn/dstate-16-plas.dat)
[Eval Data](https://paddle-org.bj.bcebos.com/paddlescience/datasets/epnn/dstress-16-plas.dat) | [Paper](https://arxiv.org/abs/2204.12088) |
+| 受力分析和逆问题 | [3D 汽车控制臂变形](./zh/examples/control_arm.md) | 机理驱动 | MLP | 无监督学习 | - | - |
| 拓扑优化 | [2D 拓扑优化](./zh/examples/topopt.md) | 数据驱动 | TopOptNN | 监督学习 | [Data](https://paddle-org.bj.bcebos.com/paddlescience/datasets/topopt/top_dataset.h5) | [Paper](https://arxiv.org/pdf/1709.09578) |
diff --git a/docs/zh/examples/control_arm.md b/docs/zh/examples/control_arm.md
new file mode 100644
index 000000000..4eece6462
--- /dev/null
+++ b/docs/zh/examples/control_arm.md
@@ -0,0 +1,521 @@
+# Control arm
+
+
+
+=== "模型训练命令"
+
+ === "正问题:受力分析求解"
+
+ ``` sh
+ # linux
+ wget https://paddle-org.bj.bcebos.com/paddlescience/datasets/control_arm/control_arm.stl -P ./datasets/
+ # windows
+ # curl https://paddle-org.bj.bcebos.com/paddlescience/datasets/control_arm/control_arm.stl --output ./datasets/control_arm.stl
+ python forward_analysis.py
+ ```
+
+ === "逆问题:参数逆推求解"
+
+ ``` sh
+ # linux
+ wget https://paddle-org.bj.bcebos.com/paddlescience/datasets/control_arm/control_arm.stl -P ./datasets/
+ # windows
+ # curl https://paddle-org.bj.bcebos.com/paddlescience/datasets/control_arm/control_arm.stl --output ./datasets/control_arm.stl
+ python inverse_parameter.py TRAIN.pretrained_model_path=https://paddle-org.bj.bcebos.com/paddlescience/models/control_arm/forward_x_axis_pretrained.pdparams
+ ```
+
+=== "模型评估命令"
+
+ === "正问题:受力分析求解"
+
+ ``` sh
+ # linux
+ wget https://paddle-org.bj.bcebos.com/paddlescience/datasets/control_arm/control_arm.stl -P ./datasets/
+ # windows
+ # curl https://paddle-org.bj.bcebos.com/paddlescience/datasets/control_arm/control_arm.stl --output ./datasets/control_arm.stl
+ python forward_analysis.py mode=eval EVAL.pretrained_model_path=https://paddle-org.bj.bcebos.com/paddlescience/models/control_arm/forward_x_axis_pretrained.pdparams
+ ```
+
+ === "逆问题:参数逆推求解"
+
+ ``` sh
+ # linux
+ wget https://paddle-org.bj.bcebos.com/paddlescience/datasets/control_arm/control_arm.stl -P ./datasets/
+ # windows
+ # curl https://paddle-org.bj.bcebos.com/paddlescience/datasets/control_arm/control_arm.stl --output ./datasets/control_arm.stl
+ python inverse_parameter.py mode=eval EVAL.pretrained_model_path=https://paddle-org.bj.bcebos.com/paddlescience/models/control_arm/inverse_x_axis_pretrained.pdparams
+ ```
+
+| 预训练模型 | 指标 |
+|:--| :--|
+| [inverse_x_axis_pretrained.pdparams](https://paddle-org.bj.bcebos.com/paddlescience/models/control_arm/inverse_x_axis_pretrained.pdparams) | loss(geo_eval): 0.02505
MAE.lambda_(geo_eval): 0.01580
MAE.mu(geo_eval): 0.01984 |
+
+## 1. 背景简介
+
+结构受力分析是在符合某个边界条件的结构受到特定条件的载荷后,结构会产生相应的应力应变,此时对它们状态的分析。 应力是一个物理量,用于描述物体内部由于外力而产生的单位面积上的力。应变则描述了物体的形状和尺寸的变化。 通常结构力学问题分为静力学问题和动力学问题,本案例着眼于静力学分析,即结构达到受力平衡状态后再进行分析。 本问题假设结构受到一个比较小的力,此时结构形变符合线弹性方程。
+
+需要指出的是,能够适用线弹性方程的结构需要满足在受力后能够完全恢复原状,即没有永久变形。 这种假设在很多情况下是合理的,但同时对于某些可能产生永久形变的材料(如塑料或橡胶)来说,这种假设可能不准确。 要全面理解形变,还需要考虑其他因素,例如物体的初始形状和尺寸、外力的历史、材料的其他物理性质(如热膨胀系数和密度)等。
+
+汽车控制臂,也称为悬挂臂或悬挂控制臂,是连接车轮和车辆底盘的重要零件。控制臂作为汽车悬架系统的导向和传力元件,将作用在车轮上的各种力传递给车身,同时保证车轮按一定轨迹运动。控制臂分别通过球铰或者衬套把车轮和车身弹性地连接在一起
+,控制臂(包括与之相连的衬套及球头)应有足够的刚度、强度和使用寿命。
+
+本问题主要研究如下汽车悬挂控制臂结构上的受力分析情况以及验证在不给定附加数据的情况下进行参数逆推的可能性,并使用深度学习方法根据线弹性等方程进行求解,结构如下所示,左侧单一圆环内表面受力,右侧两圆环内表面固定,共研究了受力方向为:x 轴负方向、z 轴正方向两种情况,下面以 x 轴正方向受力为例进行说明。
+
+
+ { loading=lazy }
+ 控制臂结构示意图
+
+
+## 2. 问题定义
+
+线弹性方程是描述物体在受力后恢复原状的能力的数学模型,表征为应力和应变之间的线性关系,其中系数被称为弹性模量(或杨氏模量),它的公式为:
+
+$$
+\begin{cases}
+ stress\_disp_{xx} = \lambda(\dfrac{\partial u}{\partial x} + \dfrac{\partial v}{\partial y} + \dfrac{\partial w}{\partial z}) + 2\mu \dfrac{\partial u}{\partial x} - \sigma_{xx} \\
+ stress\_disp_{yy} = \lambda(\dfrac{\partial u}{\partial x} + \dfrac{\partial v}{\partial y} + \dfrac{\partial w}{\partial z}) + 2\mu \dfrac{\partial v}{\partial y} - \sigma_{yy} \\
+ stress\_disp_{zz} = \lambda(\dfrac{\partial u}{\partial x} + \dfrac{\partial v}{\partial y} + \dfrac{\partial w}{\partial z}) + 2\mu \dfrac{\partial w}{\partial z} - \sigma_{zz} \\
+ traction_{x} = n_x \sigma_{xx} + n_y \sigma_{xy} + n_z \sigma_{xz} \\
+ traction_{y} = n_y \sigma_{yx} + n_y \sigma_{yy} + n_z \sigma_{yz} \\
+ traction_{z} = n_z \sigma_{zx} + n_y \sigma_{zy} + n_z \sigma_{zz} \\
+\end{cases}
+$$
+
+其中 $(x,y,z)$ 为输入的位置坐标点,$(u,v,w)$ 为对应坐标点三个维度上的应变,$(\sigma_{xx}, \sigma_{yy}, \sigma_{zz}, \sigma_{xy}, \sigma_{xz}, \sigma_{yz})$ 为 对应坐标点三个维度上的应力。
+
+结构左侧圆环内表面受到受到均匀分布的,沿 z 轴正方向大小为 $0.0025$ 的均匀应力。其它参数包括弹性模量 $E=1$,泊松比 $\nu=0.3$。目标求解该金属件表面每个点的 $u$、$v$、$w$、$\sigma_{xx}$、$\sigma_{yy}$、$\sigma_{zz}$、$\sigma_{xy}$、$\sigma_{xz}$、$\sigma_{yz}$ 共 9 个物理量。常量定义代码如下:
+
+``` yaml linenums="28"
+--8<--
+examples/control_arm/conf/forward_analysis.yaml:28:32
+--8<--
+```
+
+``` py linenums="32"
+--8<--
+examples/control_arm/forward_analysis.py:32:34
+--8<--
+```
+
+## 3. 问题求解
+
+接下来开始讲解如何将问题一步一步地转化为 PaddleScience 代码,用深度学习的方法求解该问题。
+为了快速理解 PaddleScience,接下来仅对模型构建、方程构建、计算域构建等关键步骤进行阐述,而其余细节请参考 [API文档](../api/arch.md)。
+
+### 3.1 受力分析求解
+
+#### 3.1.1 模型构建
+
+如上所述,每一个已知的坐标点 $(x, y, z)$ 都有对应的待求解的未知量:三个方向的应变 $(u, v, w)$ 和应力 $(\sigma_{xx}, \sigma_{yy}, \sigma_{zz}, \sigma_{xy}, \sigma_{xz}, \sigma_{yz})$。
+
+考虑到两组物理量对应着不同的方程,因此使用两个模型来分别预测这两组物理量:
+
+$$
+\begin{cases}
+u, v, w = f(x,y,z) \\
+\sigma_{xx}, \sigma_{yy}, \sigma_{zz}, \sigma_{xy}, \sigma_{xz}, \sigma_{yz} = g(x,y,z)
+\end{cases}
+$$
+
+上式中 $f$ 即为应变模型 `disp_net`,$g$ 为应力模型 `stress_net`,因为两者共享输入,因此在 PaddleScience 分别定义这两个网络模型后,再使用 `ppsci.arch.ModelList` 进行封装,用 PaddleScience 代码表示如下:
+
+``` py linenums="20"
+--8<--
+examples/control_arm/forward_analysis.py:20:24
+--8<--
+```
+
+#### 3.1.2 方程构建
+
+线弹性方程使用 PaddleScience 内置的 `LinearElasticity` 即可。
+
+``` py linenums="36"
+--8<--
+examples/control_arm/forward_analysis.py:36:41
+--8<--
+```
+
+#### 3.1.3 计算域构建
+
+本问题的几何区域由 stl 文件指定,按照本文档起始处"模型训练命令"下载并解压到 `./datasets/` 文件夹下。
+
+**注:数据集中的 stl 文件来自网络**。
+
+???+ warning "注意"
+
+ **使用 `Mesh` 类之前,必须先按照[1.4.3 额外依赖安装[可选]](https://paddlescience-docs.readthedocs.io/zh/latest/zh/install_setup/#143)文档,安装好 open3d、pysdf、PyMesh 3 个几何依赖包。**
+
+然后通过 PaddleScience 内置的 STL 几何类 `ppsci.geometry.Mesh` 即可读取、解析几何文件,得到计算域,并获取几何结构边界:
+
+``` py linenums="43"
+--8<--
+examples/control_arm/forward_analysis.py:43:47
+--8<--
+```
+
+#### 3.1.4 超参数设定
+
+接下来需要在配置文件中指定训练轮数,此处按实验经验,使用 2000 轮训练轮数,每轮进行 1000 步优化。
+
+``` yaml linenums="61"
+--8<--
+examples/control_arm/conf/forward_analysis.yaml:61:63
+--8<--
+```
+
+#### 3.1.5 优化器构建
+
+训练过程会调用优化器来更新模型参数,此处选择较为常用的 `Adam` 优化器,并配合使用机器学习中常用的 ExponentialDecay 学习率调整策略。
+
+``` py linenums="23"
+--8<--
+examples/control_arm/forward_analysis.py:26:30
+--8<--
+```
+
+#### 3.1.6 约束构建
+
+本问题共涉及到 4 个约束,分别为左侧圆环内表面受力的约束、右侧两圆环内表面固定的约束、结构表面边界条件的约束和结构内部点的约束。在具体约束构建之前,可以先构建数据读取配置,以便后续构建多个约束时复用该配置。
+
+``` py linenums="49"
+--8<--
+examples/control_arm/forward_analysis.py:49:59
+--8<--
+```
+
+##### 3.1.6.1 内部点约束
+
+以作用在结构内部点的 `InteriorConstraint` 为例,代码如下:
+
+``` py linenums="102"
+--8<--
+examples/control_arm/forward_analysis.py:102:138
+--8<--
+```
+
+`InteriorConstraint` 的第一个参数是方程(组)表达式,用于描述如何计算约束目标,此处填入在 [3.1.2 方程构建](#312) 章节中实例化好的 `equation["LinearElasticity"].equations`;
+
+第二个参数是约束变量的目标值,在本问题中希望与 LinearElasticity 方程相关的 9 个值 `equilibrium_x`, `equilibrium_y`, `equilibrium_z`, `stress_disp_xx`, `stress_disp_yy`, `stress_disp_zz`, `stress_disp_xy`, `stress_disp_xz`, `stress_disp_yz` 均被优化至 0;
+
+第三个参数是约束方程作用的计算域,此处填入在 [3.1.3 计算域构建](#313) 章节实例化好的 `geom["geo"]` 即可;
+
+第四个参数是在计算域上的采样配置,此处设置 `batch_size` 为:
+
+``` yaml linenums="79"
+--8<--
+examples/control_arm/conf/forward_analysis.yaml:79:79
+--8<--
+```
+
+第五个参数是损失函数,此处选用常用的 MSE 函数,且 `reduction` 设置为 `"sum"`,即会将参与计算的所有数据点产生的损失项求和;
+
+第六个参数是几何点筛选,需要对 geo 上采样出的点进行筛选,此处传入一个 lambda 筛选函数即可,其接受点集构成的张量 `x, y, z`,返回布尔值张量,表示每个点是否符合筛选条件,不符合为 `False`,符合为 `True`,因为本案例结构来源于网络,参数不完全精确,因此增加 `1e-1` 作为可容忍的采样误差。
+
+第七个参数是每个点参与损失计算时的权重,此处我们使用 `"sdf"` 表示使用每个点到边界的最短距离(符号距离函数值)来作为权重,这种 sdf 加权的方法可以加大远离边界(难样本)点的权重,减少靠近边界的(简单样本)点的权重,有利于提升模型的精度和收敛速度。
+
+第八个参数是约束条件的名字,需要给每一个约束条件命名,方便后续对其索引。此处命名为 "INTERIOR" 即可。
+
+##### 3.1.6.2 边界约束
+
+结构左侧圆环内表面受力,其上的每个点受到均匀分布的载荷,参照 [2. 问题定义](#2),大小存放在参数 $T$ 中。实际上为 x 负方向的载荷,大小为 $0.0025$,其余方向应力为 0,有如下边界条件约束:
+
+``` py linenums="62"
+--8<--
+examples/control_arm/forward_analysis.py:62:74
+--8<--
+```
+
+结构右侧两圆环内表面固定,所以其上的点在三个方向的形变均为 0,因此有如下的边界约束条件:
+
+``` py linenums="75"
+--8<--
+examples/control_arm/forward_analysis.py:75:88
+--8<--
+```
+
+结构表面不受任何载荷,即三个方向的内力平衡,合力为 0,有如下边界条件约束:
+
+``` py linenums="89"
+--8<--
+examples/control_arm/forward_analysis.py:89:101
+--8<--
+```
+
+在方程约束、边界约束构建完毕之后,以刚才的命名为关键字,封装到一个字典中,方便后续访问。
+
+``` py linenums="144"
+--8<--
+examples/control_arm/forward_analysis.py:144:150
+--8<--
+```
+
+#### 3.1.7 可视化器构建
+
+在模型评估时,如果评估结果是可以可视化的数据,可以选择合适的可视化器来对输出结果进行可视化。
+
+可视化器的输入数据通过调用 PaddleScience 的 API `sample_interior` 产生,输出数据是对应的 9 个预测的物理量,通过设置 `ppsci.visualize.VisualizerVtu` ,可以将评估的输出数据保存成 **vtu格式** 文件,最后用可视化软件打开查看即可。
+
+``` py linenums="152"
+--8<--
+examples/control_arm/forward_analysis.py:152:184
+--8<--
+```
+
+#### 3.1.8 模型训练
+
+完成上述设置之后,只需要将上述实例化的对象按顺序传递给 `ppsci.solver.Solver`,然后启动训练。
+
+``` py linenums="186"
+--8<--
+examples/control_arm/forward_analysis.py:186:208
+--8<--
+```
+
+训练后调用 `ppsci.solver.Solver.plot_loss_history` 可以将训练中的 `loss` 画出:
+
+``` py linenums="210"
+--8<--
+examples/control_arm/forward_analysis.py:210:211
+--8<--
+```
+
+另外本案例中提供了并行训练的设置,注意打开数据并行后,应该将学习率应该增大为 `原始学习率*并行卡数`,以保证训练效果。具体细节请参考 [使用指南 2.1.1 数据并行](../user_guide.md)。
+
+``` py linenums="17"
+--8<--
+examples/control_arm/forward_analysis.py:17:18
+--8<--
+```
+
+``` py linenums="140"
+--8<--
+examples/control_arm/forward_analysis.py:140:142
+--8<--
+```
+
+#### 3.1.9 模型评估与可视化
+
+训练完成或下载预训练模型后,通过本文档起始处“模型评估命令”进行模型评估和可视化。
+
+评估和可视化过程不需要进行优化器等构建,仅需构建模型、计算域、评估器(本案例不包括)、可视化器,然后按顺序传递给 `ppsci.solver.Solver` 启动评估和可视化。
+
+``` py linenums="214"
+--8<--
+examples/control_arm/forward_analysis.py:214:281
+--8<--
+```
+
+### 3.2 参数逆推求解
+
+#### 3.2.1 模型构建
+
+进行参数逆推的前提是需要知道每一个已知的坐标点 $(x, y, z)$ ,以及对应的三个方向的应变 $(u, v, w)$ 和应力 $(\sigma_{xx}, \sigma_{yy}, \sigma_{zz}, \sigma_{xy}, \sigma_{xz}, \sigma_{yz})$。这些变量的来源可以是真实数据,或数值模拟数据,或已经训练好的正问题模型。在本案例中,我们不使用任何数据,而是使用 [3.1 受力分析求解](#31) 章节中训练得到的模型来获取这些变量,因此仍然需要构建这部分模型,并为 `disp_net` 和 `stress_net` 加载正问题求解得到的权重参数作为预训练模型,注意将这两个模型冻结,以减少反向传播时间和内存占用。
+
+参数逆推中需要求解两个未知量:线弹性方程的参数 $\lambda$ 和 $\mu$,使用两个模型来分别预测这两组物理量:
+
+$$
+\begin{cases}
+\lambda = f(x,y,z) \\
+\mu = g(x,y,z)
+\end{cases}
+$$
+
+上式中 $f$ 即为求解 $\lambda$ 的模型 `inverse_lambda_net`,$g$ 为求解 $\mu$ 模型 `inverse_mu_net`。
+
+因为上述两个模型与`disp_net` 和 `stress_net` 共四个模型共享输入,因此在 PaddleScience 分别定义这四个网络模型后,再使用 `ppsci.arch.ModelList` 进行封装,用 PaddleScience 代码表示如下:
+
+``` py linenums="16"
+--8<--
+examples/control_arm/inverse_parameter.py:16:27
+--8<--
+```
+
+#### 3.2.2 方程构建
+
+线弹性方程使用 PaddleScience 内置的 `LinearElasticity` 即可。
+
+``` py linenums="35"
+--8<--
+examples/control_arm/inverse_parameter.py:35:40
+--8<--
+```
+
+#### 3.2.3 计算域构建
+
+本问题的几何区域由 stl 文件指定,按照本文档起始处"模型训练命令"下载并解压到 `./datasets/` 文件夹下。
+
+**注:数据集中的 stl 文件来自网络**。
+
+???+ warning "注意"
+
+ **使用 `Mesh` 类之前,必须先按照[1.4.3 额外依赖安装[可选]](https://paddlescience-docs.readthedocs.io/zh/latest/zh/install_setup/#143)文档,安装好 open3d、pysdf、PyMesh 3 个几何依赖包。**
+
+然后通过 PaddleScience 内置的 STL 几何类 `ppsci.geometry.Mesh` 即可读取、解析几何文件,得到计算域,并获取几何结构边界:
+
+``` py linenums="42"
+--8<--
+examples/control_arm/inverse_parameter.py:42:48
+--8<--
+```
+
+#### 3.2.4 超参数设定
+
+接下来需要在配置文件中指定训练轮数,此处按实验经验,使用 100 轮训练轮数,每轮进行 100 步优化。
+
+``` yaml linenums="73"
+--8<--
+examples/control_arm/conf/inverse_parameter.yaml:73:75
+--8<--
+```
+
+#### 3.2.5 优化器构建
+
+由于 `disp_net` 和 `stress_net` 模型的作用仅为提供三个方向的应变 $(u, v, w)$ 和应力 $(\sigma_{xx}, \sigma_{yy}, \sigma_{zz}, \sigma_{xy}, \sigma_{xz}, \sigma_{yz})$ 的值,并不需要进行训练,因此在构建优化器时需要注意不要使用 [3.2.1 模型构建](#321) 中封装的 `ModelList` 作为参数,而是使用 `inverse_lambda_net` 和 `inverse_mu_net` 组成的元组作为参数。
+
+训练过程会调用优化器来更新模型参数,此处选择较为常用的 `Adam` 优化器,并配合使用机器学习中常用的 `ExponentialDecay` 学习率调整策略。
+
+``` py linenums="29"
+--8<--
+examples/control_arm/inverse_parameter.py:29:33
+--8<--
+```
+
+#### 3.2.6 约束构建
+
+本问题共涉及到 1 个约束,为结构内部点的约束 `InteriorConstraint`。
+
+``` py linenums="50"
+--8<--
+examples/control_arm/inverse_parameter.py:50:83
+--8<--
+```
+
+`InteriorConstraint` 的第一个参数是方程(组)表达式,用于描述如何计算约束目标,此处填入在 [3.2.2 方程构建](#322) 章节中实例化好的 `equation["LinearElasticity"].equations`;
+
+第二个参数是约束变量的目标值,在本问题中希望与 LinearElasticity 方程相关且饱含参数 $\lambda$ 和 $\mu$ 的 6 个值 `stress_disp_xx`, `stress_disp_yy`, `stress_disp_zz`, `stress_disp_xy`, `stress_disp_xz`, `stress_disp_yz` 均被优化至 0;
+
+第三个参数是约束方程作用的计算域,此处填入在 [3.2.3 计算域构建](#323) 章节实例化好的 `geom["geo"]` 即可;
+
+第四个参数是在计算域上的采样配置,此处设置 `batch_size` 为:
+
+``` yaml linenums="88"
+--8<--
+examples/control_arm/conf/inverse_parameter.yaml:88:88
+--8<--
+```
+
+第五个参数是损失函数,此处选用常用的 MSE 函数,且 `reduction` 设置为 `"sum"`,即会将参与计算的所有数据点产生的损失项求和;
+
+第六个参数是几何点筛选,需要对 geo 上采样出的点进行筛选,此处传入一个 lambda 筛选函数即可,其接受点集构成的张量 `x, y, z`,返回布尔值张量,表示每个点是否符合筛选条件,不符合为 `False`,符合为 `True`;
+
+第七个参数是约束条件的名字,需要给每一个约束条件命名,方便后续对其索引。此处命名为 "INTERIOR" 即可。
+
+约束构建完毕之后,以刚才的命名为关键字,封装到一个字典中,方便后续访问。
+
+``` py linenums="84"
+--8<--
+examples/control_arm/inverse_parameter.py:84:84
+--8<--
+```
+
+#### 3.2.7 评估器构建
+
+在训练过程中通常会按一定轮数间隔,用验证集(测试集)评估当前模型的训练情况,由于我们使用正问题的预训练模型提供数据,因此已知 `label` 的值约为 $\lambda=0.57692$ 和 $\mu=0.38462$。将其包装成字典传递给 `ppsci.validate.GeometryValidator` 构造评估器并封装。
+
+``` py linenums="86"
+--8<--
+examples/control_arm/inverse_parameter.py:86:113
+--8<--
+```
+
+#### 3.2.8 可视化器构建
+
+在模型评估时,如果评估结果是可以可视化的数据,可以选择合适的可视化器来对输出结果进行可视化。
+
+可视化器的输入数据通过调用 PaddleScience 的 API `sample_interior` 产生,输出数据是$\lambda$ 和 $\mu$预测的物理量,通过设置 `ppsci.visualize.VisualizerVtu` ,可以将评估的输出数据保存成 **vtu格式** 文件,最后用可视化软件打开查看即可。
+
+``` py linenums="115"
+--8<--
+examples/control_arm/inverse_parameter.py:115:140
+--8<--
+```
+
+#### 3.2.9 模型训练
+
+完成上述设置之后,只需要将上述实例化的对象按顺序传递给 `ppsci.solver.Solver`,然后启动训练。
+
+``` py linenums="142"
+--8<--
+examples/control_arm/inverse_parameter.py:142:165
+--8<--
+```
+
+训练后调用 `ppsci.solver.Solver.plot_loss_history` 可以将训练中的 `loss` 画出:
+
+``` py linenums="167"
+--8<--
+examples/control_arm/inverse_parameter.py:167:168
+--8<--
+```
+
+#### 3.2.10 模型评估与可视化
+
+训练完成或下载预训练模型后,通过本文档起始处“模型评估命令”进行模型评估和可视化。
+
+评估和可视化过程不需要进行优化器等构建,仅需构建模型、计算域、评估器(本案例不包括)、可视化器,然后按顺序传递给 `ppsci.solver.Solver` 启动评估和可视化。
+
+``` py linenums="171"
+--8<--
+examples/control_arm/inverse_parameter.py:171:265
+--8<--
+```
+
+
+## 4. 完整代码
+
+``` py linenums="1" title="forward_analysis.py"
+--8<--
+examples/control_arm/forward_analysis.py
+--8<--
+```
+
+``` py linenums="1" title="inverse_parameter.py"
+--8<--
+examples/control_arm/inverse_parameter.py
+--8<--
+```
+
+## 5. 结果展示
+
+### 5.1 受力分析求解
+
+下面展示了当力的方向为 x 正方向时 3 个方向的应变 $u, v, w$ 以及 6 个应力 $\sigma_{xx}, \sigma_{yy}, \sigma_{zz}, \sigma_{xy}, \sigma_{xz}, \sigma_{yz}$ 的模型预测结果,结果基本符合认知。
+
+
+ 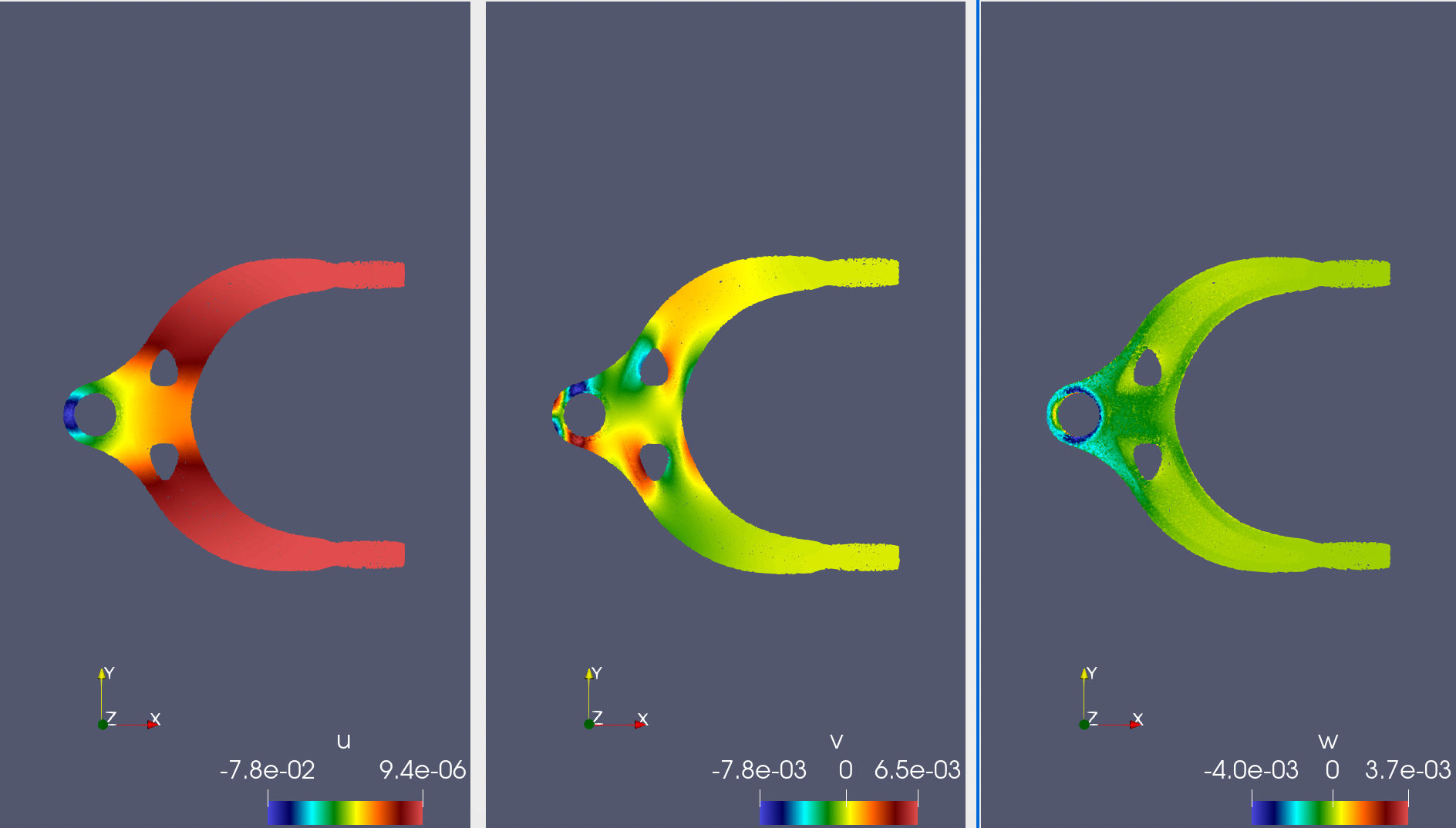{ loading=lazy }
+ 左侧为预测的结构应变 u;中间为预测的结构应变 v;右侧为预测的结构应变 w
+
+
+
+ { loading=lazy }
+ 左侧为预测的结构应力 sigma_xx;中间为预测的结构应力 sigma_xy;右侧为预测的结构应力 sigma_xz
+
+
+
+ { loading=lazy }
+ 左侧为预测的结构应力 sigma_yy;中间为预测的结构应力 sigma_yz;右侧为预测的结构应力 sigma_zz
+
+
+### 5.2 参数逆推求解
+
+下面展示了线弹性方程参数 $\lambda, \mu$ 的模型预测结果,在结构的大部分区域预测误差在 1% 左右。
+
+| data | lambda | mu |
+| :---: | :---: | :---: |
+| outs(mean) | 0.54950 | 0.38642 |
+| label | 0.57692 | 0.38462 |
+
+
+ { loading=lazy }
+ 左侧为预测的 lambda;右侧为预测的 mu
+
diff --git a/examples/control_arm/conf/forward_analysis.yaml b/examples/control_arm/conf/forward_analysis.yaml
new file mode 100644
index 000000000..2ae0aeaa9
--- /dev/null
+++ b/examples/control_arm/conf/forward_analysis.yaml
@@ -0,0 +1,89 @@
+hydra:
+ run:
+ # dynamic output directory according to running time and override name
+ dir: outputs_control_arm/forward/${now:%Y-%m-%d}/${now:%H-%M-%S}/${hydra.job.override_dirname}
+ job:
+ name: ${mode} # name of logfile
+ chdir: false # keep current working direcotry unchaned
+ config:
+ override_dirname:
+ exclude_keys:
+ - TRAIN.checkpoint_path
+ - TRAIN.pretrained_model_path
+ - EVAL.pretrained_model_path
+ - mode
+ - output_dir
+ - log_freq
+ sweep:
+ # output directory for multirun
+ dir: ${hydra.run.dir}
+ subdir: ./
+
+# general settings
+mode: train # running mode: train/eval
+seed: 2023
+output_dir: ${hydra:run.dir}
+log_freq: 100
+
+# set working condition
+NU: 0.3
+E: 1
+# T: [0, 0, 0.0025] # +Z axis
+T: [-0.0025, 0, 0] # -X axis
+
+# set geometry file path
+GEOM_PATH: ./datasets/control_arm.stl
+
+# set geometry parameter
+CIRCLE_LEFT_CENTER_XY: [-4.4, 0]
+CIRCLE_LEFT_RADIUS: 1.65
+CIRCLE_RIGHT_CENTER_XZ: [15.8, 0]
+CIRCLE_RIGHT_RADIUS: 2.21
+
+# model settings
+MODEL:
+ disp_net:
+ input_keys: ["x", "y", "z"]
+ output_keys: ["u", "v", "w"]
+ num_layers: 6
+ hidden_size: 512
+ activation: "silu"
+ weight_norm: true
+ stress_net:
+ input_keys: ["x", "y", "z"]
+ output_keys: ["sigma_xx", "sigma_yy", "sigma_zz", "sigma_xy", "sigma_xz", "sigma_yz"]
+ num_layers: 6
+ hidden_size: 512
+ activation: "silu"
+ weight_norm: true
+
+# training settings
+TRAIN:
+ epochs: 2000
+ iters_per_epoch: 1000
+ save_freq: 100
+ eval_freq: 100
+ eval_during_train: false
+ eval_with_no_grad: false
+ lr_scheduler:
+ epochs: ${TRAIN.epochs}
+ iters_per_epoch: ${TRAIN.iters_per_epoch}
+ learning_rate: 1.0e-4
+ gamma: 0.95
+ decay_steps: 15000
+ by_epoch: false
+ batch_size:
+ arm_left: 128
+ arm_right: 256
+ arm_surface: 4096
+ arm_interior: 2048
+ visualizer_vtu: 100000
+ weight:
+ arm_right: {"u": 1, "v": 1, "w": 1}
+ pretrained_model_path: null
+ checkpoint_path: null
+
+# evaluation settings
+EVAL:
+ eval_with_no_grad: true
+ pretrained_model_path: null
diff --git a/examples/control_arm/conf/inverse_parameter.yaml b/examples/control_arm/conf/inverse_parameter.yaml
new file mode 100644
index 000000000..d1eb16e9e
--- /dev/null
+++ b/examples/control_arm/conf/inverse_parameter.yaml
@@ -0,0 +1,101 @@
+hydra:
+ run:
+ # dynamic output directory according to running time and override name
+ dir: outputs_control_arm/inverse/${now:%Y-%m-%d}/${now:%H-%M-%S}/${hydra.job.override_dirname}
+ job:
+ name: ${mode} # name of logfile
+ chdir: false # keep current working direcotry unchaned
+ config:
+ override_dirname:
+ exclude_keys:
+ - TRAIN.checkpoint_path
+ - TRAIN.pretrained_model_path
+ - EVAL.pretrained_model_path
+ - mode
+ - output_dir
+ - log_freq
+ sweep:
+ # output directory for multirun
+ dir: ${hydra.run.dir}
+ subdir: ./
+
+# general settings
+mode: train # running mode: train/eval
+seed: 2023
+output_dir: ${hydra:run.dir}
+log_freq: 100
+
+# set working condition
+NU: 0.3
+E: 1
+
+# set geometry file path
+GEOM_PATH: ./datasets/control_arm.stl
+
+# set geometry parameter
+CIRCLE_LEFT_CENTER_XY: [-4.4, 0]
+CIRCLE_LEFT_RADIUS: 1.65
+CIRCLE_RIGHT_CENTER_XZ: [15.8, 0]
+CIRCLE_RIGHT_RADIUS: 2.21
+
+# model settings
+MODEL:
+ disp_net:
+ input_keys: ["x", "y", "z"]
+ output_keys: ["u", "v", "w"]
+ num_layers: 6
+ hidden_size: 512
+ activation: "silu"
+ weight_norm: true
+ stress_net:
+ input_keys: ["x", "y", "z"]
+ output_keys: ["sigma_xx", "sigma_yy", "sigma_zz", "sigma_xy", "sigma_xz", "sigma_yz"]
+ num_layers: 6
+ hidden_size: 512
+ activation: "silu"
+ weight_norm: true
+ inverse_lambda_net:
+ input_keys: ["x", "y", "z"]
+ output_keys: ["lambda_",]
+ num_layers: 6
+ hidden_size: 512
+ activation: "silu"
+ weight_norm: true
+ inverse_mu_net:
+ input_keys: ["x", "y", "z"]
+ output_keys: ["mu",]
+ num_layers: 6
+ hidden_size: 512
+ activation: "silu"
+ weight_norm: true
+
+# training settings
+TRAIN:
+ epochs: 100
+ iters_per_epoch: 100
+ save_freq: 1
+ eval_freq: 1
+ eval_during_train: true
+ eval_with_no_grad: false
+ lr_scheduler:
+ epochs: ${TRAIN.epochs}
+ iters_per_epoch: ${TRAIN.iters_per_epoch}
+ learning_rate: 1.0e-4
+ gamma: 0.95
+ decay_steps: 100
+ by_epoch: false
+ batch_size:
+ arm_interior: 2000
+ visualizer_vtu: 100000
+ pretrained_model_path: null
+ checkpoint_path: null
+
+# evaluation settings
+EVAL:
+ pretrained_model_path: null
+ eval_with_no_grad: true
+ total_size:
+ validator: 20000
+ batch_size:
+ validator: 1024
+ visualizer_vtu: 100000
diff --git a/examples/control_arm/forward_analysis.py b/examples/control_arm/forward_analysis.py
new file mode 100644
index 000000000..c8fe365be
--- /dev/null
+++ b/examples/control_arm/forward_analysis.py
@@ -0,0 +1,297 @@
+from os import path as osp
+
+import hydra
+import numpy as np
+from omegaconf import DictConfig
+from paddle import distributed as dist
+
+import ppsci
+from ppsci.utils import logger
+
+
+def train(cfg: DictConfig):
+ # set random seed for reproducibility
+ ppsci.utils.misc.set_random_seed(cfg.seed)
+ # initialize logger
+ logger.init_logger("ppsci", osp.join(cfg.output_dir, f"{cfg.mode}.log"), "info")
+ # set parallel
+ enable_parallel = dist.get_world_size() > 1
+
+ # set model
+ disp_net = ppsci.arch.MLP(**cfg.MODEL.disp_net)
+ stress_net = ppsci.arch.MLP(**cfg.MODEL.stress_net)
+ # wrap to a model_list
+ model_list = ppsci.arch.ModelList((disp_net, stress_net))
+
+ # set optimizer
+ lr_scheduler = ppsci.optimizer.lr_scheduler.ExponentialDecay(
+ **cfg.TRAIN.lr_scheduler
+ )()
+ optimizer = ppsci.optimizer.Adam(lr_scheduler)(model_list)
+
+ # specify parameters
+ LAMBDA_ = cfg.NU * cfg.E / ((1 + cfg.NU) * (1 - 2 * cfg.NU))
+ MU = cfg.E / (2 * (1 + cfg.NU))
+
+ # set equation
+ equation = {
+ "LinearElasticity": ppsci.equation.LinearElasticity(
+ E=None, nu=None, lambda_=LAMBDA_, mu=MU, dim=3
+ )
+ }
+
+ # set geometry
+ control_arm = ppsci.geometry.Mesh(cfg.GEOM_PATH)
+ geom = {"geo": control_arm}
+ # set bounds
+ BOUNDS_X, BOUNDS_Y, BOUNDS_Z = control_arm.bounds
+
+ # set dataloader config
+ train_dataloader_cfg = {
+ "dataset": "NamedArrayDataset",
+ "iters_per_epoch": cfg.TRAIN.iters_per_epoch,
+ "sampler": {
+ "name": "BatchSampler",
+ "drop_last": True,
+ "shuffle": True,
+ },
+ "num_workers": 1,
+ }
+
+ # set constraint
+ arm_left_constraint = ppsci.constraint.BoundaryConstraint(
+ equation["LinearElasticity"].equations,
+ {"traction_x": cfg.T[0], "traction_y": cfg.T[1], "traction_z": cfg.T[2]},
+ geom["geo"],
+ {**train_dataloader_cfg, "batch_size": cfg.TRAIN.batch_size.arm_left},
+ ppsci.loss.MSELoss("sum"),
+ criteria=lambda x, y, z: np.sqrt(
+ np.square(x - cfg.CIRCLE_LEFT_CENTER_XY[0])
+ + np.square(y - cfg.CIRCLE_LEFT_CENTER_XY[1])
+ )
+ <= cfg.CIRCLE_LEFT_RADIUS + 1e-1,
+ name="BC_LEFT",
+ )
+ arm_right_constraint = ppsci.constraint.BoundaryConstraint(
+ {"u": lambda d: d["u"], "v": lambda d: d["v"], "w": lambda d: d["w"]},
+ {"u": 0, "v": 0, "w": 0},
+ geom["geo"],
+ {**train_dataloader_cfg, "batch_size": cfg.TRAIN.batch_size.arm_right},
+ ppsci.loss.MSELoss("sum"),
+ criteria=lambda x, y, z: np.sqrt(
+ np.square(x - cfg.CIRCLE_RIGHT_CENTER_XZ[0])
+ + np.square(z - cfg.CIRCLE_RIGHT_CENTER_XZ[1])
+ )
+ <= cfg.CIRCLE_RIGHT_RADIUS + 1e-1,
+ weight_dict=cfg.TRAIN.weight.arm_right,
+ name="BC_RIGHT",
+ )
+ arm_surface_constraint = ppsci.constraint.BoundaryConstraint(
+ equation["LinearElasticity"].equations,
+ {"traction_x": 0, "traction_y": 0, "traction_z": 0},
+ geom["geo"],
+ {**train_dataloader_cfg, "batch_size": cfg.TRAIN.batch_size.arm_surface},
+ ppsci.loss.MSELoss("sum"),
+ criteria=lambda x, y, z: np.sqrt(
+ np.square(x - cfg.CIRCLE_LEFT_CENTER_XY[0])
+ + np.square(y - cfg.CIRCLE_LEFT_CENTER_XY[1])
+ )
+ > cfg.CIRCLE_LEFT_RADIUS + 1e-1,
+ name="BC_SURFACE",
+ )
+ arm_interior_constraint = ppsci.constraint.InteriorConstraint(
+ equation["LinearElasticity"].equations,
+ {
+ "equilibrium_x": 0,
+ "equilibrium_y": 0,
+ "equilibrium_z": 0,
+ "stress_disp_xx": 0,
+ "stress_disp_yy": 0,
+ "stress_disp_zz": 0,
+ "stress_disp_xy": 0,
+ "stress_disp_xz": 0,
+ "stress_disp_yz": 0,
+ },
+ geom["geo"],
+ {**train_dataloader_cfg, "batch_size": cfg.TRAIN.batch_size.arm_interior},
+ ppsci.loss.MSELoss("sum"),
+ criteria=lambda x, y, z: (
+ (BOUNDS_X[0] < x)
+ & (x < BOUNDS_X[1])
+ & (BOUNDS_Y[0] < y)
+ & (y < BOUNDS_Y[1])
+ & (BOUNDS_Z[0] < z)
+ & (z < BOUNDS_Z[1])
+ ),
+ weight_dict={
+ "equilibrium_x": "sdf",
+ "equilibrium_y": "sdf",
+ "equilibrium_z": "sdf",
+ "stress_disp_xx": "sdf",
+ "stress_disp_yy": "sdf",
+ "stress_disp_zz": "sdf",
+ "stress_disp_xy": "sdf",
+ "stress_disp_xz": "sdf",
+ "stress_disp_yz": "sdf",
+ },
+ name="INTERIOR",
+ )
+
+ # re-assign to cfg.TRAIN.iters_per_epoch
+ if enable_parallel:
+ cfg.TRAIN.iters_per_epoch = len(arm_left_constraint.data_loader)
+
+ # wrap constraints togetherg
+ constraint = {
+ arm_left_constraint.name: arm_left_constraint,
+ arm_right_constraint.name: arm_right_constraint,
+ arm_surface_constraint.name: arm_surface_constraint,
+ arm_interior_constraint.name: arm_interior_constraint,
+ }
+
+ # set visualizer(optional)
+ # add inferencer data
+ samples = geom["geo"].sample_interior(
+ cfg.TRAIN.batch_size.visualizer_vtu,
+ criteria=lambda x, y, z: (
+ (BOUNDS_X[0] < x)
+ & (x < BOUNDS_X[1])
+ & (BOUNDS_Y[0] < y)
+ & (y < BOUNDS_Y[1])
+ & (BOUNDS_Z[0] < z)
+ & (z < BOUNDS_Z[1])
+ ),
+ )
+ pred_input_dict = {
+ k: v for k, v in samples.items() if k in cfg.MODEL.disp_net.input_keys
+ }
+ visualizer = {
+ "visulzie_u_v_w_sigmas": ppsci.visualize.VisualizerVtu(
+ pred_input_dict,
+ {
+ "u": lambda out: out["u"],
+ "v": lambda out: out["v"],
+ "w": lambda out: out["w"],
+ "sigma_xx": lambda out: out["sigma_xx"],
+ "sigma_yy": lambda out: out["sigma_yy"],
+ "sigma_zz": lambda out: out["sigma_zz"],
+ "sigma_xy": lambda out: out["sigma_xy"],
+ "sigma_xz": lambda out: out["sigma_xz"],
+ "sigma_yz": lambda out: out["sigma_yz"],
+ },
+ prefix="vis",
+ )
+ }
+
+ # initialize solver
+ solver = ppsci.solver.Solver(
+ model_list,
+ constraint,
+ cfg.output_dir,
+ optimizer,
+ lr_scheduler,
+ cfg.TRAIN.epochs,
+ cfg.TRAIN.iters_per_epoch,
+ seed=cfg.seed,
+ equation=equation,
+ geom=geom,
+ save_freq=cfg.TRAIN.save_freq,
+ log_freq=cfg.log_freq,
+ eval_freq=cfg.TRAIN.eval_freq,
+ eval_during_train=cfg.TRAIN.eval_during_train,
+ eval_with_no_grad=cfg.TRAIN.eval_with_no_grad,
+ visualizer=visualizer,
+ checkpoint_path=cfg.TRAIN.checkpoint_path,
+ )
+
+ # train model
+ solver.train()
+
+ # plot losses
+ solver.plot_loss_history(by_epoch=True, smooth_step=1)
+
+
+def evaluate(cfg: DictConfig):
+ # set random seed for reproducibility
+ ppsci.utils.misc.set_random_seed(cfg.seed)
+ # initialize logger
+ logger.init_logger("ppsci", osp.join(cfg.output_dir, f"{cfg.mode}.log"), "info")
+
+ # set model
+ disp_net = ppsci.arch.MLP(**cfg.MODEL.disp_net)
+ stress_net = ppsci.arch.MLP(**cfg.MODEL.stress_net)
+ # wrap to a model_list
+ model_list = ppsci.arch.ModelList((disp_net, stress_net))
+
+ # set geometry
+ control_arm = ppsci.geometry.Mesh(cfg.GEOM_PATH)
+ # geometry bool operation
+ geo = control_arm
+ geom = {"geo": geo}
+ # set bounds
+ BOUNDS_X, BOUNDS_Y, BOUNDS_Z = control_arm.bounds
+
+ # set visualizer(optional)
+ # add inferencer data
+ samples = geom["geo"].sample_interior(
+ cfg.TRAIN.batch_size.visualizer_vtu,
+ criteria=lambda x, y, z: (
+ (BOUNDS_X[0] < x)
+ & (x < BOUNDS_X[1])
+ & (BOUNDS_Y[0] < y)
+ & (y < BOUNDS_Y[1])
+ & (BOUNDS_Z[0] < z)
+ & (z < BOUNDS_Z[1])
+ ),
+ )
+ pred_input_dict = {
+ k: v for k, v in samples.items() if k in cfg.MODEL.disp_net.input_keys
+ }
+ visualizer = {
+ "visulzie_u_v_w_sigmas": ppsci.visualize.VisualizerVtu(
+ pred_input_dict,
+ {
+ "u": lambda out: out["u"],
+ "v": lambda out: out["v"],
+ "w": lambda out: out["w"],
+ "sigma_xx": lambda out: out["sigma_xx"],
+ "sigma_yy": lambda out: out["sigma_yy"],
+ "sigma_zz": lambda out: out["sigma_zz"],
+ "sigma_xy": lambda out: out["sigma_xy"],
+ "sigma_xz": lambda out: out["sigma_xz"],
+ "sigma_yz": lambda out: out["sigma_yz"],
+ },
+ prefix="vis",
+ )
+ }
+
+ # initialize solver
+ solver = ppsci.solver.Solver(
+ model_list,
+ output_dir=cfg.output_dir,
+ seed=cfg.seed,
+ geom=geom,
+ log_freq=cfg.log_freq,
+ eval_with_no_grad=cfg.EVAL.eval_with_no_grad,
+ visualizer=visualizer,
+ pretrained_model_path=cfg.EVAL.pretrained_model_path,
+ )
+
+ # visualize prediction after finished training
+ solver.visualize()
+
+
+@hydra.main(
+ version_base=None, config_path="./conf", config_name="forward_analysis.yaml"
+)
+def main(cfg: DictConfig):
+ if cfg.mode == "train":
+ train(cfg)
+ elif cfg.mode == "eval":
+ evaluate(cfg)
+ else:
+ raise ValueError(f"cfg.mode should in ['train', 'eval'], but got '{cfg.mode}'")
+
+
+if __name__ == "__main__":
+ main()
diff --git a/examples/control_arm/inverse_parameter.py b/examples/control_arm/inverse_parameter.py
new file mode 100644
index 000000000..473f27fa5
--- /dev/null
+++ b/examples/control_arm/inverse_parameter.py
@@ -0,0 +1,281 @@
+from os import path as osp
+
+import hydra
+from omegaconf import DictConfig
+
+import ppsci
+from ppsci.utils import logger
+
+
+def train(cfg: DictConfig):
+ # set random seed for reproducibility
+ ppsci.utils.misc.set_random_seed(cfg.seed)
+ # initialize logger
+ logger.init_logger("ppsci", osp.join(cfg.output_dir, f"{cfg.mode}.log"), "info")
+
+ # set model
+ disp_net = ppsci.arch.MLP(**cfg.MODEL.disp_net)
+ stress_net = ppsci.arch.MLP(**cfg.MODEL.stress_net)
+ inverse_lambda_net = ppsci.arch.MLP(**cfg.MODEL.inverse_lambda_net)
+ inverse_mu_net = ppsci.arch.MLP(**cfg.MODEL.inverse_mu_net)
+ # freeze models
+ disp_net.freeze()
+ stress_net.freeze()
+ # wrap to a model_list
+ model = ppsci.arch.ModelList(
+ (disp_net, stress_net, inverse_lambda_net, inverse_mu_net)
+ )
+
+ # set optimizer
+ lr_scheduler = ppsci.optimizer.lr_scheduler.ExponentialDecay(
+ **cfg.TRAIN.lr_scheduler
+ )()
+ optimizer = ppsci.optimizer.Adam(lr_scheduler)((inverse_lambda_net, inverse_mu_net))
+
+ # set equation
+ equation = {
+ "LinearElasticity": ppsci.equation.LinearElasticity(
+ E=None, nu=None, lambda_="lambda_", mu="mu", dim=3
+ )
+ }
+
+ # set geometry
+ control_arm = ppsci.geometry.Mesh(cfg.GEOM_PATH)
+ # geometry bool operation
+ geo = control_arm
+ geom = {"geo": geo}
+ # set bounds
+ BOUNDS_X, BOUNDS_Y, BOUNDS_Z = control_arm.bounds
+
+ # set dataloader config
+ interior_constraint = ppsci.constraint.InteriorConstraint(
+ equation["LinearElasticity"].equations,
+ {
+ "stress_disp_xx": 0,
+ "stress_disp_yy": 0,
+ "stress_disp_zz": 0,
+ "stress_disp_xy": 0,
+ "stress_disp_xz": 0,
+ "stress_disp_yz": 0,
+ },
+ geom["geo"],
+ {
+ "dataset": "NamedArrayDataset",
+ "iters_per_epoch": cfg.TRAIN.iters_per_epoch,
+ "sampler": {
+ "name": "BatchSampler",
+ "drop_last": True,
+ "shuffle": True,
+ },
+ "num_workers": 1,
+ "batch_size": cfg.TRAIN.batch_size.arm_interior,
+ },
+ ppsci.loss.MSELoss("sum"),
+ criteria=lambda x, y, z: (
+ (BOUNDS_X[0] < x)
+ & (x < BOUNDS_X[1])
+ & (BOUNDS_Y[0] < y)
+ & (y < BOUNDS_Y[1])
+ & (BOUNDS_Z[0] < z)
+ & (z < BOUNDS_Z[1])
+ ),
+ name="INTERIOR",
+ )
+ constraint = {interior_constraint.name: interior_constraint}
+
+ # set validator
+ LAMBDA_ = cfg.NU * cfg.E / ((1 + cfg.NU) * (1 - 2 * cfg.NU)) # 0.5769
+ MU = cfg.E / (2 * (1 + cfg.NU)) # 0.3846
+ geom_validator = ppsci.validate.GeometryValidator(
+ {
+ "lambda_": lambda out: out["lambda_"],
+ "mu": lambda out: out["mu"],
+ },
+ {
+ "lambda_": LAMBDA_,
+ "mu": MU,
+ },
+ geom["geo"],
+ {
+ "dataset": "NamedArrayDataset",
+ "sampler": {
+ "name": "BatchSampler",
+ "drop_last": False,
+ "shuffle": False,
+ },
+ "total_size": cfg.EVAL.total_size.validator,
+ "batch_size": cfg.EVAL.batch_size.validator,
+ },
+ ppsci.loss.MSELoss("sum"),
+ metric={"MAE": ppsci.metric.MAE()},
+ name="geo_eval",
+ )
+ validator = {geom_validator.name: geom_validator}
+
+ # set visualizer(optional)
+ # add inferencer data
+ samples = geom["geo"].sample_interior(
+ cfg.TRAIN.batch_size.visualizer_vtu,
+ criteria=lambda x, y, z: (
+ (BOUNDS_X[0] < x)
+ & (x < BOUNDS_X[1])
+ & (BOUNDS_Y[0] < y)
+ & (y < BOUNDS_Y[1])
+ & (BOUNDS_Z[0] < z)
+ & (z < BOUNDS_Z[1])
+ ),
+ )
+ pred_input_dict = {
+ k: v for k, v in samples.items() if k in cfg.MODEL.disp_net.input_keys
+ }
+ visualizer = {
+ "visulzie_lambda_mu": ppsci.visualize.VisualizerVtu(
+ pred_input_dict,
+ {
+ "lambda": lambda out: out["lambda_"],
+ "mu": lambda out: out["mu"],
+ },
+ prefix="vis",
+ )
+ }
+
+ # initialize solver
+ solver = ppsci.solver.Solver(
+ model,
+ constraint,
+ cfg.output_dir,
+ optimizer,
+ lr_scheduler,
+ cfg.TRAIN.epochs,
+ cfg.TRAIN.iters_per_epoch,
+ seed=cfg.seed,
+ equation=equation,
+ geom=geom,
+ save_freq=cfg.TRAIN.save_freq,
+ log_freq=cfg.log_freq,
+ eval_freq=cfg.TRAIN.eval_freq,
+ eval_during_train=cfg.TRAIN.eval_during_train,
+ eval_with_no_grad=cfg.TRAIN.eval_with_no_grad,
+ validator=validator,
+ visualizer=visualizer,
+ pretrained_model_path=cfg.TRAIN.pretrained_model_path,
+ )
+
+ # train model
+ solver.train()
+
+ # plot losses
+ solver.plot_loss_history(by_epoch=False, smooth_step=1, use_semilogy=True)
+
+
+def evaluate(cfg: DictConfig):
+ # set random seed for reproducibility
+ ppsci.utils.misc.set_random_seed(cfg.seed)
+ # initialize logger
+ logger.init_logger("ppsci", osp.join(cfg.output_dir, f"{cfg.mode}.log"), "info")
+
+ # set model
+ disp_net = ppsci.arch.MLP(**cfg.MODEL.disp_net)
+ stress_net = ppsci.arch.MLP(**cfg.MODEL.stress_net)
+ inverse_lambda_net = ppsci.arch.MLP(**cfg.MODEL.inverse_lambda_net)
+ inverse_mu_net = ppsci.arch.MLP(**cfg.MODEL.inverse_mu_net)
+ # wrap to a model_list
+ model = ppsci.arch.ModelList(
+ (disp_net, stress_net, inverse_lambda_net, inverse_mu_net)
+ )
+
+ # set geometry
+ control_arm = ppsci.geometry.Mesh(cfg.GEOM_PATH)
+ # geometry bool operation
+ geo = control_arm
+ geom = {"geo": geo}
+ # set bounds
+ BOUNDS_X, BOUNDS_Y, BOUNDS_Z = control_arm.bounds
+
+ # set validator
+ LAMBDA_ = cfg.NU * cfg.E / ((1 + cfg.NU) * (1 - 2 * cfg.NU)) # 0.57692
+ MU = cfg.E / (2 * (1 + cfg.NU)) # 0.38462
+ geom_validator = ppsci.validate.GeometryValidator(
+ {
+ "lambda_": lambda out: out["lambda_"],
+ "mu": lambda out: out["mu"],
+ },
+ {
+ "lambda_": LAMBDA_,
+ "mu": MU,
+ },
+ geom["geo"],
+ {
+ "dataset": "NamedArrayDataset",
+ "sampler": {
+ "name": "BatchSampler",
+ "drop_last": False,
+ "shuffle": False,
+ },
+ "total_size": cfg.EVAL.total_size.validator,
+ "batch_size": cfg.EVAL.batch_size.validator,
+ },
+ ppsci.loss.MSELoss("sum"),
+ metric={"MAE": ppsci.metric.MAE()},
+ name="geo_eval",
+ )
+ validator = {geom_validator.name: geom_validator}
+
+ # set visualizer(optional)
+ # add inferencer data
+ samples = geom["geo"].sample_interior(
+ cfg.EVAL.batch_size.visualizer_vtu,
+ criteria=lambda x, y, z: (
+ (BOUNDS_X[0] < x)
+ & (x < BOUNDS_X[1])
+ & (BOUNDS_Y[0] < y)
+ & (y < BOUNDS_Y[1])
+ & (BOUNDS_Z[0] < z)
+ & (z < BOUNDS_Z[1])
+ ),
+ )
+ pred_input_dict = {
+ k: v for k, v in samples.items() if k in cfg.MODEL.disp_net.input_keys
+ }
+ visualizer = {
+ "visulzie_lambda_mu": ppsci.visualize.VisualizerVtu(
+ pred_input_dict,
+ {
+ "lambda": lambda out: out["lambda_"],
+ "mu": lambda out: out["mu"],
+ },
+ prefix="vis",
+ )
+ }
+
+ # initialize solver
+ solver = ppsci.solver.Solver(
+ model,
+ output_dir=cfg.output_dir,
+ seed=cfg.seed,
+ log_freq=cfg.log_freq,
+ eval_with_no_grad=cfg.EVAL.eval_with_no_grad,
+ validator=validator,
+ visualizer=visualizer,
+ pretrained_model_path=cfg.EVAL.pretrained_model_path,
+ )
+ # evaluate after finished training
+ solver.eval()
+ # visualize prediction after finished training
+ solver.visualize()
+
+
+@hydra.main(
+ version_base=None, config_path="./conf", config_name="inverse_parameter.yaml"
+)
+def main(cfg: DictConfig):
+ if cfg.mode == "train":
+ train(cfg)
+ elif cfg.mode == "eval":
+ evaluate(cfg)
+ else:
+ raise ValueError(f"cfg.mode should in ['train', 'eval'], but got '{cfg.mode}'")
+
+
+if __name__ == "__main__":
+ main()
diff --git a/mkdocs.yml b/mkdocs.yml
index fed9fb88d..18088a48a 100644
--- a/mkdocs.yml
+++ b/mkdocs.yml
@@ -64,6 +64,7 @@ nav:
- 结构:
- Biharmonic2D: zh/examples/biharmonic2d.md
- Bracket: zh/examples/bracket.md
+ - Control_arm: zh/examples/control_arm.md
- EPNN: zh/examples/epnn.md
- Phy-LSTM: zh/examples/phylstm.md
- TopOpt: zh/examples/topopt.md
diff --git a/ppsci/arch/base.py b/ppsci/arch/base.py
index 5d173cf00..32b6c7ac4 100644
--- a/ppsci/arch/base.py
+++ b/ppsci/arch/base.py
@@ -124,6 +124,20 @@ def register_output_transform(
"""
self._output_transform = transform
+ def freeze(self):
+ """Freeze all parameters."""
+ for param in self.named_parameters():
+ param.stop_gradient = True
+
+ self.eval()
+
+ def unfreeze(self):
+ """Unfreeze all parameters."""
+ for param in self.named_parameters():
+ param.stop_gradient = False
+
+ self.train()
+
def __str__(self):
num_fc = 0
num_conv = 0