A Python framework for Probabilistic Acoustic Sediment Mapping. See project website here for more details
Software Developer: Dr. Daniel Buscombe, Northern Arizona University, Flagstaff, AZ 86011, Now at Marda Science, LLC.
Example backscatter data (.tiff files) originate from data collected by R2Sonic and distributed for use as part of the R2Sonic 2017 Multispectral Backscatter competition.
Bed observation data from Patricia Bay are digitized from data presented in: B.~Biffard. Seabed remote sensing by single-beam echosounder: models, methods and applications. Doctoral dissertation, University of Victoria, Canada, 2011.
Bed observation data from Portsmouth (NEWBEX) are digitized from data presented in: T.~Weber, L.~and Ward. Observations of backscatter from sand and gravel seafloors between 170 and 250 kHz. Journal of the Acoustical Society of America, vol.~138, no.~4, pp.~2169 - 2180, 2015.
CRF subfunctions using the pydensecrf wrapper
PriSM is designed for use with Python 3
conda env create --file env/prism.yml
conda activate prism
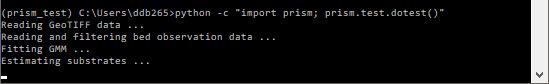
Then run the test ::
python -c "import prism; prism.test.dotest()"
run the GUI ::
python -c "import prism; prism.gui_funcs.gui()"
or alternatively from within the python console like so:
Download the user manual
A full worked example using the NEWBEX data set
def run_prism():
#==================================================================================
##GMM parameters
covariance = 'full'
tol = 1e-2
##CRF parameters
theta = 300
mu = 100
n_iter = 15
# general settings
gridres = 1 # grid size in m
buff = 10 # buffer distance on each bed observation
prob_thres = 0.1 #probability threshold
chambolle = 0.0 #chambolle filter
test_size = 0.5
## update this with the full file path
bs100 = 'newbex_mosaic_100.tiff'
bs200 = 'newbex_mosaic_200.tiff'
bs400 = 'newbex_mosaic_400.tiff'
refs_file = 'newbex_bed.shp'
prefix = 'newbex'
#======== READ
#==================================================================================
input = [bs100, bs200, bs400]
img, bs = read_geotiff(input, gridres, chambolle)
bed = read_shpfile(refs_file, bs)
Lc = get_sparse_labels(bs, bed, buff)
if np.ndim(img)>2:
mask = img[:,:,0]==0
else:
mask = img==0
#======== GMM
#==================================================================================
g = fit_GMM(img, Lc, test_size, covariance, tol)
y_pred_gmm, y_prob_gmm, y_gmm_prob_per_class = apply_GMM(g, img, prob_thres)
#======== CRF
#==================================================================================
y_pred_crf, y_prob_crf, y_crf_prob_per_class = apply_CRF(img, Lc, bed['labels'], n_iter, prob_thres, theta, mu)
#======== PLOT
#==================================================================================
cmap = plt.get_cmap('tab20b',len(bed['labels'])-1).colors
cmap1 = []
cmap1.append('gray')
for k in cmap:
cmap1.append(colors.rgb2hex(k))
plot_dists_per_sed(Lc, img, bed, cmap1, prefix)
plot_gmm(mask, y_pred_gmm, y_prob_gmm, bs, bed, cmap, prefix)
plot_crf(mask, y_pred_crf, y_prob_crf, bs, bed, cmap, prefix)
plot_gmm_image(mask, y_pred_gmm, y_prob_gmm, bs, bed, cmap, prefix)
plot_crf_image(mask, y_pred_crf, y_prob_crf, bs, bed, cmap, prefix)
plot_gmm_crf(mask, y_pred_gmm, y_prob_gmm, y_pred_crf, y_prob_crf, bs, bed, cmap, prefix)
plot_gmm_crf_images(mask, y_pred_gmm, y_prob_gmm, y_pred_crf, y_prob_crf, bs, bed, cmap, prefix)
plot_bs_maps(img, bed, bs, cmap, prefix)
plot_confmatCRF(y_pred_crf, Lc, bed, prefix)
plot_confmatGMM(y_pred_gmm, Lc, bed, prefix)
#======== EXPORT
#==================================================================================
export_bed_data(bed, prefix)
export_gmm_gtiff(mask, y_pred_gmm.copy(), y_prob_gmm.copy(), bs, prefix)
export_crf_gtiff(mask, y_pred_crf.copy(), y_prob_crf.copy(), bs, prefix)
if __name__ == '__main__':
run_prism()
v. 0.1. 2/26/2018. Initial public release 11/16/21 minor upgrade / test - still works!