diff --git a/NMDS.png b/NMDS.png
new file mode 100644
index 0000000..be1f87b
Binary files /dev/null and b/NMDS.png differ
diff --git a/README.md b/README.md
index dd23da0..3a3fa1a 100644
--- a/README.md
+++ b/README.md
@@ -1,14 +1,28 @@
# Initial NMDS ordinations and example diversity statistics for metabarcoding data
-## This workflow and Jupyter binder shows a basic workflow to visualize metabarcoding data in R.
+#### This workflow and Jupyter binder shows a basic workflow to visualize metabarcoding data in R.
+
+### Quick Start:
+#### Click on the binder link, than click "knit" to make an html R Markdown. You may need to turn off your pop up blocker.
+
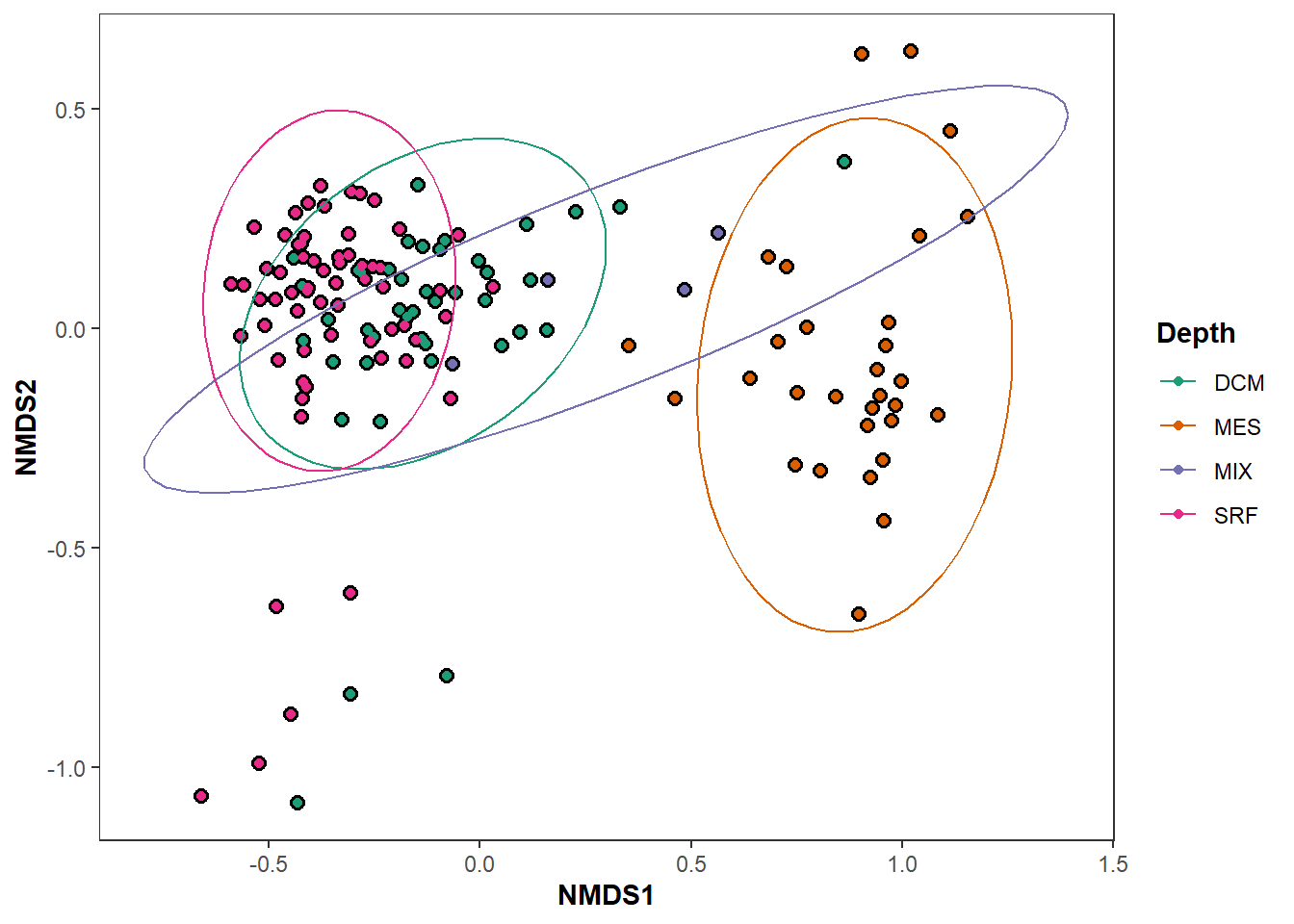
+### Example Output:
+
+
+### Summary:
+#### This workflow will generate several example NMDS ordinations from an OTU/ASV table.
+#### Please remove taxonomic assignments before using this workflow.
+#### This plot will provide a visual representation of a given sample’s community composition in relation to all other samples. These plots allow you to quickly visualize your metabarcoding community by any metadata grouping (categorical) factor you choose.
+
### Authors:
-### Jason Rothman
+#### Jason Rothman
-### Jane Lucas
+#### Jane Lucas
### Links
#### GitHub: https://github.com/jasonarothman/Metabarcoding_viz
-#### Binder: [](https://mybinder.org/v2/gh/jasonarothman/Metabarcoding_viz/master?filepath=rstudio)
\ No newline at end of file
+#### Binder: [](https://mybinder.org/v2/gh/jasonarothman/Metabarcoding_viz/master?urlpath=rstudio)
+
+### We hope this is helpful!
\ No newline at end of file
diff --git a/install.R b/install.R
index 6ae6ad4..410f429 100644
--- a/install.R
+++ b/install.R
@@ -1,2 +1,4 @@
-pkgs = c("ggplot2", "rmarkdown", "vegan", "RColorBrewer")
-install.packages(pkgs)
\ No newline at end of file
+install.packages("ggplot2")
+install.packages("rmarkdown")
+install.packages("vegan")
+install.packages("RColorBrewer")
\ No newline at end of file
 https://orcid.org/0000-0002-4848-8901
https://orcid.org/0000-0002-4848-8901