This repository has been archived by the owner on Jan 13, 2021. It is now read-only.
-
Notifications
You must be signed in to change notification settings - Fork 31
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
- Loading branch information
Adam M. Wilson
committed
Oct 2, 2018
1 parent
389f6a4
commit af47864
Showing
54 changed files
with
2,726 additions
and
6 deletions.
There are no files selected for viewing
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,189 @@ | ||
| --- | ||
| title: "Raster Analyses" | ||
| --- | ||
|
|
||
| ```{r echo=F, eval=T, message=F,warning=F} | ||
| library(sf) | ||
| library(spData) | ||
| library(viridis) | ||
| library(tidyverse) | ||
| library(raster) | ||
| ``` | ||
|
|
||
| --- | ||
|
|
||
| # Spatial Data in R | ||
|
|
||
| ## Available Packages | ||
|
|
||
| * `sp` First major spatial data package/format | ||
| * `rgdal` reading and writing spatial data | ||
| * `rgeos` Interface to open-source geometry engine (GEOS) | ||
| * `sf` Spatial Features in the 'tidyverse' | ||
| * `raster` gridded data (like satellite imagery) | ||
| * and a few others... | ||
|
|
||
| ## Common Simple Feature (SF) types | ||
|
|
||
| 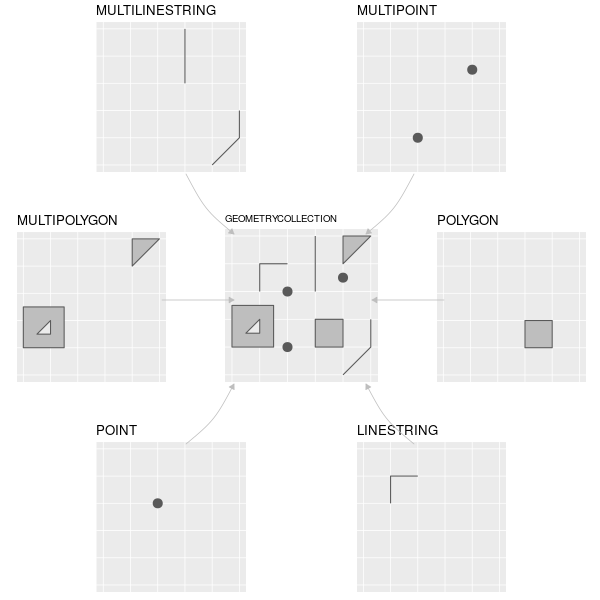 | ||
|
|
||
| ## What about grids? | ||
| ```{r echo=F, message=F,warning=F} | ||
| library(raster) | ||
| library(spData) | ||
| data(elev) #load fake data from spData package | ||
| plot(elev) | ||
| ``` | ||
|
|
||
|
|
||
| # Raster Data | ||
|
|
||
| ## Raster Data Types | ||
| Many types including: | ||
|
|
||
| * Thematic (classified/thematic) data: land-use or soils data. | ||
| * Continuous data (temperature, elevation, spectral) | ||
| * Imagery / Pictures (scanned maps, drawings, etc.) | ||
|
|
||
| Most Common: | ||
|
|
||
| * Satellite Imagery and derived products | ||
| * Earth System Models (e.g. climate models) | ||
| * Other models | ||
|
|
||
|
|
||
| ## Raster data in the tidyverse | ||
|
|
||
| Raster data is not yet closely connected to the **tidyverse**, however: | ||
|
|
||
| - Some functions from `raster` work well in `pipes` | ||
| - Convert vector data to `Spatial*` form using `as(my_vector, "Spatial")` for raster-vector interactions | ||
| - Some early efforts to bring raster data into the **tidyverse**, including [tabularaster](https://github.com/hypertidy/tabularaster), [sfraster](https://github.com/mdsumner/sfraster), [fasterize](https://github.com/ecohealthalliance/fasterize), and [stars](https://github.com/r-spatial/stars) (multidimensional, large datasets). | ||
|
|
||
| ## Raster Package | ||
| ```{r message=F,warning=F} | ||
| library(raster) | ||
| library(spData) | ||
| data(elev) #load fake data from spData package | ||
| plot(elev) #load fake data from spData package | ||
| ``` | ||
|
|
||
|
|
||
|
|
||
| ## Raster Data Structure | ||
| ```{r} | ||
| elev | ||
| ``` | ||
|
|
||
| --- | ||
| ```{r} | ||
| str(elev) | ||
| ``` | ||
|
|
||
| --- | ||
|
|
||
| ### Arrays with metadata | ||
| ```{r} | ||
| as.array(elev) | ||
| ``` | ||
| Rasters are just arrays / matricies with metadata. | ||
|
|
||
| --- | ||
|
|
||
| ### Cell Stats | ||
|
|
||
| ```{r} | ||
| cellStats(elev, stat = mean, na.rm=T) | ||
| cellStats(elev, stat = quantile, na.rm=T) | ||
| ``` | ||
|
|
||
| --- | ||
|
|
||
| ### Raster Data Memory | ||
| ```{r} | ||
| inMemory(elev) | ||
| canProcessInMemory(elev) | ||
| ``` | ||
| Raster does not try to load or work with large datasets in RAM. It creates temporary files and processes them in the background. | ||
|
|
||
| ## Map Algebra | ||
|
|
||
| Convert feet to meters | ||
| ```{r} | ||
| elev_m <- elev*0.3048 | ||
| plot(elev_m) | ||
| ``` | ||
|
|
||
| --- | ||
|
|
||
| ### Simple Filter | ||
| ```{r} | ||
| high_ground <- elev_m>8 | ||
| plot(high_ground) | ||
| ``` | ||
|
|
||
|
|
||
| --- | ||
|
|
||
| ### Focal | ||
| ```{r} | ||
| m <- matrix(1,nrow=3,ncol=3) | ||
| m | ||
| elev_smooth <- focal(elev_m,m,mean) | ||
| ``` | ||
| --- | ||
| ```{r} | ||
| par(mfrow=c(1,2)) | ||
| plot(elev_m) | ||
| plot(elev_smooth) | ||
| ``` | ||
|
|
||
| ## Arbitrary functions | ||
|
|
||
| ```{r} | ||
| NA16=function(x) ifelse(x == 16,1,NA) | ||
| one_cell <- calc(elev,fun = NA16) | ||
| plot(one_cell) | ||
| ``` | ||
|
|
||
| ## Distances to non-NA cells | ||
| ```{r} | ||
| distance(one_cell)%>% | ||
| plot() | ||
| ``` | ||
|
|
||
| Distance unit is meters if RasterLayer is `+proj=longlat`, map units (typically also meters) otherwise. | ||
|
|
||
| ## And much more | ||
|
|
||
| * aggregate (to coarser resolution) | ||
| * area (calculate cell area) | ||
| * buffer | ||
| * contour | ||
| * crop | ||
| * interpolate | ||
| * Moran's I | ||
|
|
||
|
|
||
| # Visualization | ||
|
|
||
| ## ggplot | ||
|
|
||
| ```{r, message=F} | ||
| library(rasterVis) | ||
| gplot(elev)+ | ||
| geom_raster(aes(fill=value)) | ||
| ``` | ||
|
|
||
| --- | ||
|
|
||
| ### All ggplot functionality available | ||
|
|
||
| ```{r} | ||
| gplot(elev)+ | ||
| geom_raster(aes(fill=value))+ | ||
| coord_equal()+ | ||
| scale_fill_viridis_c() | ||
| ``` | ||
|
|
||
|
|
Oops, something went wrong.