Description of the approach : https://blog.goodaudience.com/heartbeat-classification-detecting-abnormal-heartbeats-and-heart-diseases-from-ecgs-913449c2665
Requirement : Keras, tensorflow, numpy
ECGs
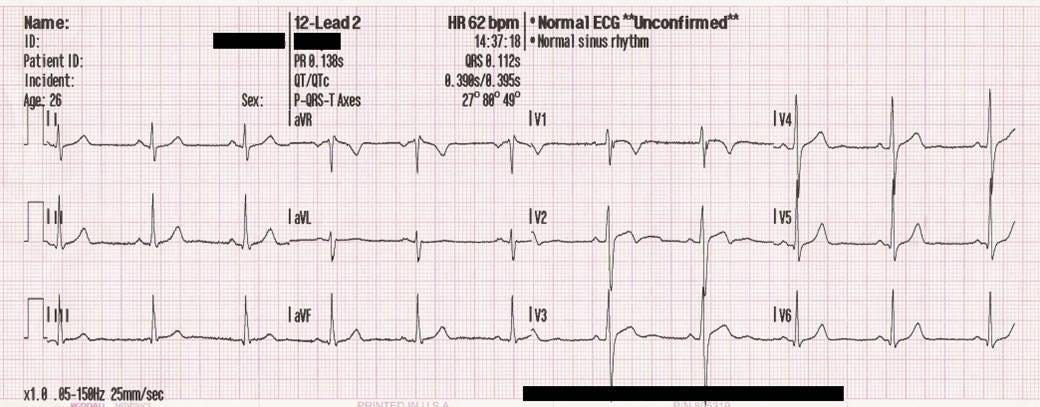
An ECG is a 1D signal that is the result of recording the electrical activity of the heart using an electrode. It is one of the tool that cardiologists use to diagnose heart anomalies and diseases.
In this blog post we are going to use an annotated dataset of heartbeats already preprocessed by the authors of this paper to see if we can train a model to detect abnormal heartbeats.
The original datasets used are the MIT-BIH Arrhythmia Dataset and The PTB Diagnostic ECG Database that were preprocessed by [1] based on the methodology described in III.A of the paper in order to end up with samples of a single heartbeat each and normalized amplitudes as :
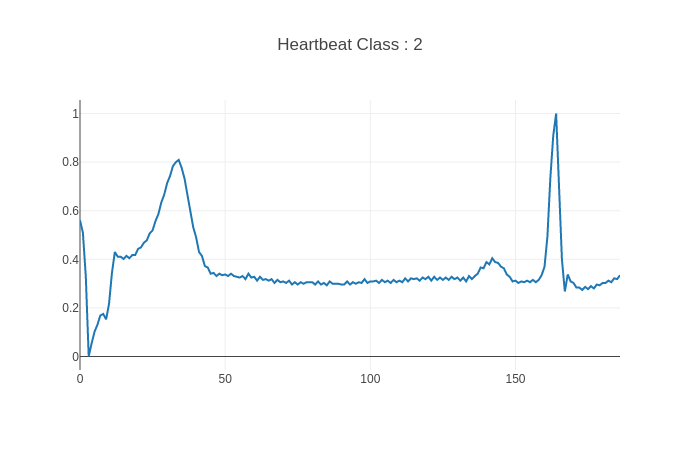
MIT-BIH Arrhythmia dataset :
- Number of Categories: 5
- Number of Samples: 109446
- Sampling Frequency: 125Hz
- Data Source: Physionet’s MIT-BIH Arrhythmia Dataset
- Classes: [’N’: 0, ‘S’: 1, ‘V’: 2, ‘F’: 3, ‘Q’: 4]
The PTB Diagnostic ECG Database
- Number of Samples: 14552
- Number of Categories: 2 ( Normal vs Abnomal)
- Sampling Frequency: 125Hz
- Data Source: Physionet’s PTB Diagnostic Database
The published preprocessed version of the MIT-BIH dataset does not fit the description that authors provided of it in their paper as the former is heavily unbalanced while the latter is not. This made it so my results are not directly comparable to theirs. I sent the authors an email to have the same split as them and I’ll update my results if I get a reply. A similar issue exists for the PTB dataset.
Similar to [1] I use a neural network based on 1D convolutions but without the residual blocks :
Figure 3 : Keras model
Code :
MIT-BIH Arrhythmia dataset :
- Accuracy : 98.5
- F1 score : 91.5
The PTB Diagnostic ECG Database
- Accuracy : 98.3
- F1 score : 98.8
Since the PTB dataset is much smaller than the MIT-BIH dataset we can try and see if the representations learned from MIT-BIH dataset can generalize and be useful to the PTB dataset and improve the performance.
This can be done by loading the weights learned in MIT-BIH as initial point of training the PTB model.
From Scratch :
- Accuracy : 98.3
- F1 score :** 98.8**
Freezing the Convolution Layer and Training the Fully connected ones :
- Accuracy : 95.6
- F1 score : 96.9
Training all layers :
- Accuracy : 99.2
- F1 score : 99.4
We can see the freezing the first layers does not work very well. But if we initialize the weights with those learned on MIT-BIH and train all layers we are able to improve the performance compared to training from scratch.
Code to reproduce the results is available at : https://github.com/CVxTz/ECG_Heartbeat_Classification