This is the repository for pymolschortucts.py. This file contains 215 functions in 25 categories mapped to short names that work like aliases (the table below is incomplete). These shortcuts include many functions that make working in PyMOL more productive and fun! Some shortcuts save the many hours of work required to assemble a new script file while other shortcuts save only a few minutes but lower motivational barriers. You do not need to understand Python to be able to run the script.
A paper about this project was published in Protien Science in January 2020. Access to the PDF is open; you do not need a subscription to Protein Science to download it.
To cite the software, use the following citation if you use BibTeX.
@Article{Mooers2020ShortcutsForFasterImageCreationInPyMOL,
author = {Mooers, Blaine HM},
title = {Shortcuts for faster image creation in PyMOL},
number = {1},
pages = {268--276},
volume = {29},
file = {:Mooers2020ShortcutsForFasterImageCreationInPyMOL.pdf:PDF},
url = {https://onlinelibrary.wiley.com/doi/pdfdirect/10.1002/pro.3781},
journal = {Protein Science},
publisher = {Wiley Online Library},
doi = {10.1002/pro.3781},
year = {2020},
}The Google Scholar generated ENDNOTE file (scholar. new) is above with the other files.
To get a quick overview of this project, scroll down through this README.md file and watch the animations. To learn more, see the PDFs in the slideshow folder:
- mooers23jul2019ACAPyMOLshortcuts.pdf (20-minute talk at the American Crystallographic Association annual meeting in Covington, KY)
- mooers18oct2019RNAcornbeltPyMOLShortcuts4RNA.pdf (9-minute talk at the Cornbelt RNA meeting at the U of Missouri in Columbia, Missouri)
- mooers26Oct2019SWTCCPyMOLshortcuts.pdf (20-minute talk, Southwest Theoretical and Computational Chemistry (SWTCC) annual meeting at the U of Oklahoma in Norman, OK)
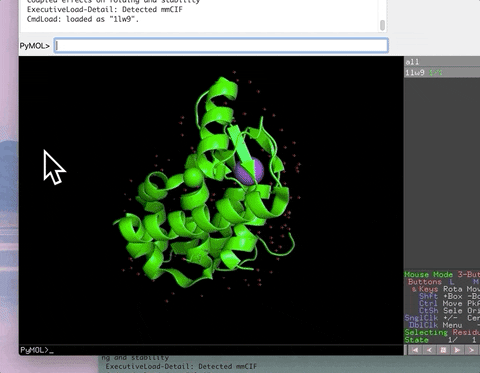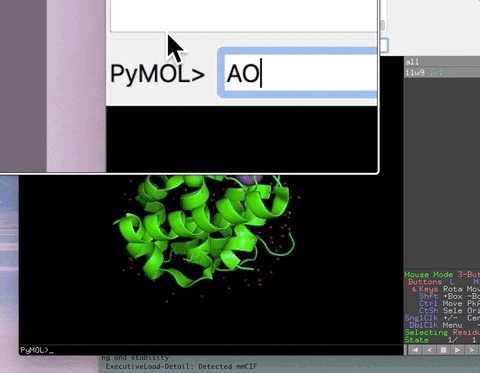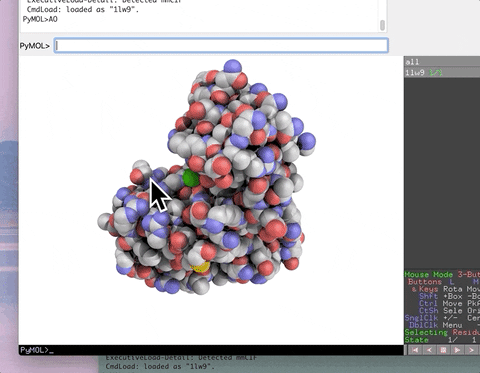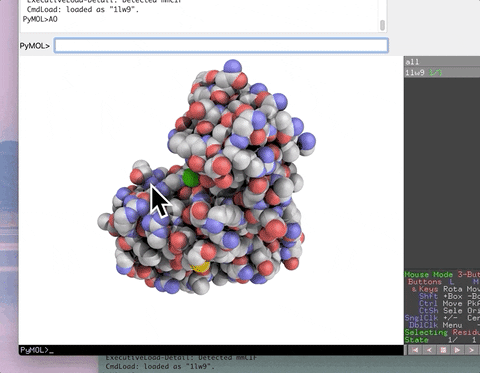
The animation below demonstrates the photorealistic effect from the ambient occlusion shortcut applied to a protein by entering two letters, AO. This shortcut executes 16 lines of code with the entry of two letters!
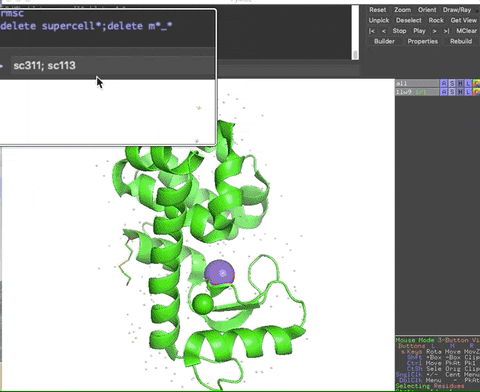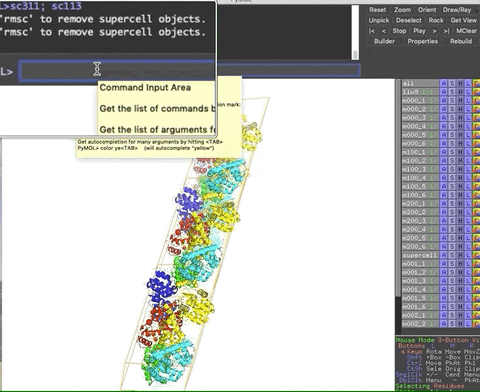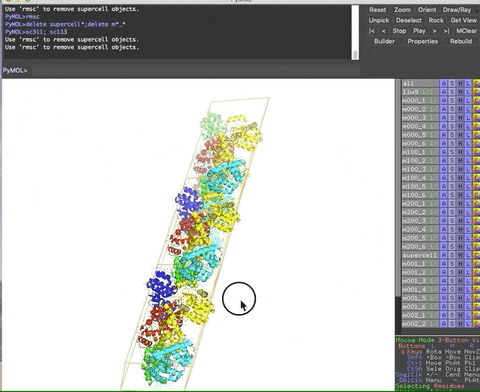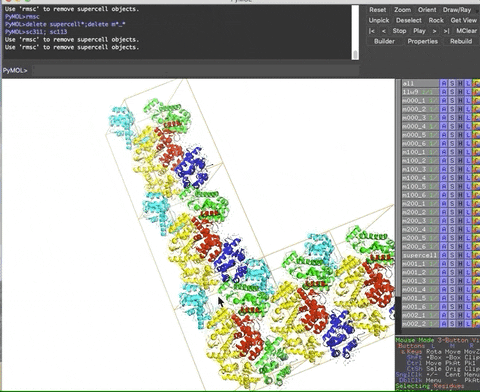
The animation below demonstrates the submission of two shortcuts, sc311 and sc113, separated by a semicolon on the command line. They generate symmetry mates that fill five unit cells. The unit cells form an L shape. Each symmetry mate appears as a separate object in the menu in the panel to the right of the vv. These can be removed by entering the shortcut rmsc.
The variant file pymolschortuctsNobs4.py lacks a few functions that depend on the Python module beautifulsoup4 that is not shipped with PyMOL. Choose this variant file if you do not know how to add beautifulsoup4 to your version of PyMOL.
If you want to use the shortcuts with a version of PyMOL that has a Python interpreter to which you can add an external module (generally any Open source PyMOL or the incentive version equal to or greater than 2.0), download the pymolshortcuts.py script.
If you have an incentive version less than 2.0 or you do not want to bother with the installation of the datetime module, download the pymolshortcutsNoDateTime.py. This version of the script lacks the functions that depend on the datetime module. By using this version You will not get error messages about this module not being found.
The Python program pip is available for all installing external modules. The recommended way of using pip when more than one Python interpreter is on our computer is to import it as a module on the command line with the path of the python interpreter for which you are making the install. For example, to install the datetime module in the macports Python3.7, use sudo -H /opt/local/bin/python3.7 -m pip install --upgrade datetime. The minus m flag means import the following module. Depending on your configuration of macports, you may not need to use sudo -H.
Before you download the script file, be sure that you have set your system to show file extensions. Both Mac and Windows operating system hide the file extension by default.
On the Mac, do the following
- Open a new Finder window.
- In the menu bar, select Finder > Preferences.
- Check the box Show all filename extension
On Windows, do the following:
- Search for File Explorer Options in the Start search box and open the Folder Options box.
- Uncheck the box next to Hide extensions for known file types".
To download all of the files, git clone the repository.
Otherwise, left-click on one of the files above and left-click "Raw".
The raw file will be displayed in your browser.
Select save under the file pull-down of your browser.
Your browser may add .txt after .py.
Delete .txt by backspacing over it.
Then save the file to your home directory.
Your browser may try to append the .py with .txt again via a popup GUI, but do not select this option.
To have the shortcuts always available in PyMOL, add the command 'run ~/pymolshortcutsNobs4.py' (recommended for most users) or 'run ~/pymolshortcutsbs4.py' (for advanced users) to the .pymolrc, pymolrc or pymolrc.pml file in your home directory. This command will to load the functions in the script on every startup of PyMOL so you do not have to think about doing so again.
The pymolrc file is an optional file. You may have to create it with a text editor if you have not done so already. If you do not have a text editor, you can use PyMOL's built-in text editor. Go to File --> Edit pymolrc. The functions will be loaded into memory but will not be executed despite the use of the command run. You may want to store the script pymolshortcuts.py in a safer place than your home directory. I store mine in /Users/blaine/Scripts/PyMOLScripts/.
A number of shortcuts open external programs. The commands that work on the Mac OS are active by default. These are not path-dependent. They should work regardless of the nonconventional location of your applications, as long as they are in your PATH. They are found around line 500 in the pymolshortcuts.py script file.
The commands for Mac OS need to be commented out and the commands for Linux or Windows need to be uncommented. Do not use a word processor to edit this file. Visual Studio Code is an excellent, free, and intuitive text editor that is platform independent. Install one of the Python in-plugins via the marketplace to get syntax highlighting of the pymolshortcuts.py script file
Around line 500, there are also paths to local directories of PDB files and webmail sites that need to be customized.
Note that multiple versions of PyMOL can be on one computer. On the Mac, you can give the package in Applications a different name. I append the version number to the stem of the file name, e.g., PyMOL233.app for version 2.3.3. Avoid trouble down the road by NOT introducing whitespaces into the file name.
The incentive version of PyMOL is the easiest to install due to the availability of installers. You can use the incentive version for 30 days for free without buying an annual license.
See the instructions on the PyMOL Wiki page for installing on the Mac.
There are many ways of installing PyMOL on the Mac. I suggest using the install command below instead for installing pymol on the Mac with Macports. I have used Fink, Anaconda, Homebrew, and Macports. I have the fewest problems with Macports, followed by Fink.
The main barrier to using Macports is the need to install the Xcode-related command-line tools. The protocol for doing so varies with the version of the Mac operating system and the version of Xcode installed on your computer. Expect to spend at least an hour installing the command line tools and then Macports. If this sounds like too much trouble, download the 30-day trial of the incentive version.
Open source version of pymol via Macports works fine. It is missing a few minor thrills found in the incentive version like the ability to import background images into the viewport.
You can have your favorite protein rocking over a scene from your last vacation.
This is cool not essential for serious work.
Note that the installation of PyMOL via MacPorts on the Mac (sudo port install pymol +python37) allows specification of the version of the Python interpreter.
PyMOL can be installed with versions 2.7, 3.5, 3.6, and 3.7 of Python.
This is handy if you have a module written for only Python3.5 that you want to import into PyMOL to do something unique and creative without writing a new plugin for PyMOL.
However, only one version of Macports PyMOL can be active at a time.
You could just run sudo port install pymol +pythonXX whenever you want to switch versions of pymol because the installation goes very quickly.
The proper way to switch ports is to enter sudo activate .
Of course, you have to have the corresponding Macports Python interpreter installed prior to installing pymol.
Open source version of pymol via Macports works fine in my hands. It has only a few minor frills missing that the incentive has.
See the PyMOL Wiki page for Linux Installs. The install protocol varies with the flavor of Linux. Install protocols for seven flavors of Linux are listed on the PyMOL Wiki. I have had success with installing recent versions of PyMOL on Ubuntu and Centos 7. The webpage also describes installing PyMOL from source code. From my past experience, this is more successful on Linux than on MacOS.
See the PyMOL Wiki page for installing PyMOL on Windows. The protocol varies with the flavor of Linux. I have had success with installing recent versions of PyMOL on Ubuntu and Centos 7.
If all of the above is overwhelmning, you do have a 30-day free-trial period with which you can use the incentive version of PyMOL.
Enter SC at the upper PyMOL> prompt to get a list of shortcuts printed to the command history window. Use the help function to see the documentation for each shortcut. For example, enter help PW to print the documentation for the shortcut PW to the command history window. The documentation has four sections:
- a description of what the shortcut does
- an example of running the shortcut,
- the corresponding pml code with one command per line for easy reuse in a script file
- all of the commands on a single line for re-use on the command line as a horizontal script.
The shortcut PW takes one or more search terms and then sends them to the PyMOL Wiki. A browser tab opens for each search term, so multiple searches are run in parallel while you continue your work in PyMOL. You can inspect the results of the searches when there is a natural break in your work in PyMOL. Other search functions can submit parallel searches of PubMed, Google, bioRxiv, Research Gate, GitHub, and more. See the tables below.
Another class of shortcuts launches your favorite full-featured text editor from within PyMOL. You have to install the text editor and edit the file path to the executable.
Another class of shortcuts opens the manuscript or grant application that you are working on in Overleaf. You have to edit the script by pasting in the appropriate link to the specific document.
Another class of shortcuts saves files with timestamps embedded in the filename to avoid overwriting png, pdb, pse, and other types of files written out from PyMOL. These save function names begin with s, (e.g., spse filename saves the current session with date and time to nearest second embedded in the file name). You can delete the unwanted version(s) at a later time. These functions are useful if you do not have these files under version control.
| Shortcut | Short Description |
|---|---|
| cntccp4s | Count number of *.ccp4 (electron density map) files in the current directory. |
| cntfiles | Count the number of files in the current directory. |
| cntlogs | Count the number of *.log files in the current directory. |
| cntmtzs | Count the number of *.mtz (structure factor) files in the current directory. |
| cntpdbs | Count the number of pdb files in the current directory. |
| cntpmls | Count the number of pml (Pymol macro language) files in the current directory. |
| cntpngs | Count the number of *.png image files in the current directory. |
| cntpses | Count the number of *.pse (session) files in the current directory. |
| Shortcut | Short Description |
|---|---|
| JASP | Open JASP from within PyMOL. |
| JMP | Open the JMP from within PyMOL. |
| RStudio | Open Rstudio GUI. |
| cranR | Open Cran R from within PyMOL. |
| ddb | Open DBBrowserSQLite. |
| Excel | Open Excel from within PyMOL. |
| jabref | Open the jabref from within PyMOL. |
| Julia | Open the Julia from within PyMOL. |
| juliapro | Open the juliapro from within PyMOL. |
| oc | Open the data analysis program octave (open source analog of matlab) from within PyMOL. |
| ppt | Open the PowerPoint from within PyMOL. |
| Shortcut | Short Description |
|---|---|
| hb | Creates an object of all H-bonds found by PyMOL. |
| rmhb | Delete all H-bonds in the selection, which is all by default. |
| Shortcut | Short Description |
|---|---|
| gimp | Open the image editing program gimp from within PyMOL. |
| inkscape | Open the image editing program inkscape from within PyMOL. |
| Shortcut | Short Description |
|---|---|
| nmr | Show all models in a nmr structure. |
| nmroff | Hide all but the first model in a nmr structure. |
| rmd | Remove all measurement objects in the interal GUI. |
| rmsc | Remove supercell objects. |
| sc111 | Make a lattice of 1 x 1 x 1 unit cells. |
| sc112 | Make a lattice of 1 x 1 x 2 unit cells |
| sc113 | Make a lattice of 1 x 1 x 3 unit cells. |
| sc121 | Make a lattice of 1 x 2 x 1 unit cells. |
| sc122 | Make a lattice of 1 x 2 x 2 unit cells. |
| sc123 | Make a lattice of 1 x 2 x 3 unit cells. |
| sc131 | Make a lattice of 1 x 3 x 1 unit cells. |
| sc132 | Make a lattice of 1 x 3 x 2 unit cells. |
| sc133 | Make a lattice of 1 x 3 x 3 unit cells. |
| sc211 | Make a lattice of 2 x 1 x 1 unit cells. |
| sc212 | Make a lattice of 2 x 1 x 2 unit cells. |
| sc213 | Make a lattice of 2 x 1 x 3 unit cells. |
| sc221 | Make a lattice of 2 x 2 x 1 unit cells. |
| sc222 | Make a lattice of 2 x 2 x 2 unit cells |
| sc231 | Make a lattice of 2 x 3 x 1 unit cells. |
| sc311 | Make a lattice of 3 x 1 x 1 unit cells. |
| sc312 | Make a lattice of 3 x 1 x 2 unit cells. |
| sc313 | Make a lattice of 3 x 1 x 3 unit cells. |
| sc321 | Make a lattice of 3 x 2 x 1 unit cells. |
| sc331 | Make a lattice of 3 x 3 x 1 unit cells. |
| sc333 | Make a lattice of 3 x 3 x 3 unit cells. |
| Shortcut | Short Description |
|---|---|
| ccp4mg | Open ccp4mg from within PyMOL. |
| chimera | Open Chimera from within PyMOL. |
| coot | Open coot from within PyMOL. |
| jmol | Open Jmol from within PyMOL. |
| vmd | Open vmd from within PyMOL. |
| yasara | Open the molecular graphics program YASASRA from within PyMOL. |
| Shortcut | Short Description |
|---|---|
| AO | Commands to make ambient occlusion image like those in Qutemole. |
| AOBW | Commands to make ambient occlusion image like those in Qutemole but coloring with grayscale. |
| AOD | Make ambient occlusion image of any with dark carbon atoms. |
| AODBW | Make ambient occlusion image of any with dark carbon atoms in grayscale. |
| BU | Commands to make the biological unit. |
| BW | Make black-and white-ribbon cartoon on a white background. |
| CB | Runs Jared Sampson's script "colorblindfriendly.py". |
| CBSS | Apply colorblind-friendly coloring to ribbon or cartoon representations. |
| CR | Commands to make colored filled-ring cartoon of nucleic acids. |
| CSS | Commands to color ribbon or cartoon representations of proteins by secondary structure. |
| DU | Make dumbbell (ribbons with rolled edges) cartoon of the main chains of nucleic acids and proteins. |
| FR | Make filled-ring cartoon of nucleic acids. |
| HH | Hide hydrogen atoms of currently visible molecular objects. |
| PE | Apply pearl effect about selection cation or anion. |
| PE125 | Apply alternative pearl effect about selection. |
| PE25 | Apply alternative pearl effect about selected cation or anion. |
| PE33 | Apply alternative pearl effect about selection. |
| PE50 | Apply alternative pearl effect about selected cation or anion. |
| PE66 | Apply alternative pearl effect about selection. |
| PE75 | Apply alternative pearl effect about selection. |
| PE85 | Apply alternative pearl effect about selection. |
| SH | Show hydrogen atoms of currently visible molecular objects. |
| bs | bs creates a ball and stick representation of an object. |
| bsbw | bs creates a gray-scaled ball-and-stick representation of an object. |
| bsbwsc | bsbwsc creates a gray-scaled ball-and-stick representation of an object or selection. |
| Only the side chains are shown as ball and stick when used with a cartoon (ribbon diagram). | |
| bstvdw | Transparent vdw surface over ball and stick representation by Robert Paton at Colorato State University. |
| cartoonbw | Grayscale by secondary structure. |
| gscale | Apply grayscale to all atoms by element. |
| rgb | Restore rgb coloring of atoms by element. |
| tvdw | Transparent vdw surface by Robert Paton at Colorado State University. |
| tvdwbw | Transparent vdw surface by Robert Paton at Colorado State University and grayscale by gscale function. |
| Shortcut | Short Description |
|---|---|
| colorh1 | Color protein molecules according to the Eisenberg hydrophobicity scale. Uses scheme 1. |
| colorh2 | Color protein molecules according to the Eisenberg hydrophobicity scale. Uses scheme 2. |
| timcolor | Tim Mather's biophysical coloring scheme for proteins. |
| yrb | A script to highlight hydrophobicity and charge on protein surfaces (Hagemans et al. 2015). |
| Shortcut | Short Description |
|---|---|
| pdbremarks | Read REMARK lines from PDB file. |
| Return dictionary with remarkNum as key and list of lines as value. | |
| Called by the function quat(). | |
| quat | Runs Thomas Holder's quat.py script to generate a biological unit using crystallographic symmetry. |
| Requires a pdb file. The defult file type with the fetch command is *.cif. | |
| When using fetch, add the optional parameter type=pdb. | |
| Of course, you can also use type=pdb1 to retrieve the biological unit form the PDB. | |
Reads REMARK 350 from the pdb file filename and creates the biological unit (quaternary structure). |
|
| quat350 | Get transformation matrices for biomolecule 1 from REMARK 350. |
| Shortcut | Short Description |
|---|---|
| buriedW | Return a selection of buried waters. |
| checkParams | Checks user params for the findSeq function. |
| interface | Returns a selection of interface residues named according to what you passed into selName. |
| Shortcut | Short Description |
|---|---|
| omx | Align long axis of molecule along the x-axis of the viewport. |
| omxy | Align long axis of molecule along minus x-y axis. |
| omxyz | Align long axis of the selection along the minu xyz axis. |
| omy | Align long axis of the selection along the y-axis of the viewport in the negative direction. |
| omz | Align long axis of selection along the z-axis of the viewport in the negative z direction. |
| ox | Align long axis of molecule along x-axis. |
| oxy | Align long axis of the selection along the x-y axis. |
| oxyz | Align long axis of the selection along the xyz axis. |
| oy | Align long axis of the selection along the y-axis of the viewport. |
| oz | Align long axis of selection along the z-axis of the viewport. |
| Shortcut | Short Description |
|---|---|
| pairD | Find the pairwise distances between two selections. |
| Shortcut | Short Description |
|---|---|
| saln | Save a aln file (alignment file) with a time stamp. |
| sccp4 | Save a ccp4 electron density map with a time stamp. |
| scif | Save a cif file (Crystallographic Information File) with a time stamp. |
| sdae | Save a dae file (Collada File) with a time stamp. |
| sdat | Save a dat file (output data file) with a time stamp. |
| sfasta | Save a fasta file (output data file) with a time stamp. |
| sidtf | Save a idtf file (Intermediate Data Text Format) with a time stamp. |
| smae | Save a mae (Maestro) file with a time stamp. |
| smmd | Save a mmd (Macromodel) file with a time stamp. |
| smmod | Save a mmod (Macromodel) file with a time stamp. |
| smoe | Save a moe file (Molecular Operating Environment) with a time stamp. |
| smol | Save a mol file with a time stamp. |
| smol2 | Save a mol2 file with a time stamp. |
| smtl | Save mtl (Wavefront Material) file format with a time stamp. |
| sobj | Save obj file (Wavefront mesh file format) with a time stamp. |
| sout | Save output data file with a time stamp. |
| spdb | Save pdb data file with a time stamp. |
| spkl | Save a pkl file (Python pickle file) with a time stamp. |
| spkla | Save a pkla file (Python pickle file) with a time stamp. |
| spmo | Save a pmo file (XYZ, binary format file) with a time stamp. |
| spng | Save a png file (Python pickle file) with a time stamp. |
| spov | Save pov (POV-ray tracing file format) file with a time stamp. |
| spqr | Save pqr with file with timestamp. |
| spse | Save session file with a time stamp. |
| srv | Get the view settings in a compact format on one line. Save to file with timestamp appended to the stem of the filename. |
| ssdf | Save session file with a time stamp. |
| swrl | Save wrl (VRML 2 file format) file with a time stamp. |
| Shortcut | Short Description |
|---|---|
| gitAdd | Enter help(gitAdd) to print steps for adding a file to version control. |
| gitCommit | Enter help(gitCommit) to print steps for saving updates to a file under version control. |
| gitInit | Enter help(gitInit) to print steps for creating a git repository. |
| gitPull | Enter help(gitPull) to print steps to update a repository on github.com. |
| gitPush | Enter help(gitPush) to print steps update a repository on github.com. |
| rline | Prints cheat sheet for the readline commands. |
| rv | Get the view settings in a compact format on one line. |
| Shortcut | Short Description |
|---|---|
| AB | Send search term or phrase to Amazon.com Books in default browser. |
| AC | Send search term to Anaconda Cloud. |
| AX | Send search term or phrase to arXiv. |
| BX | Send search term or phrase to bioRxiv |
| GB | Send search term or phrase to Google Books in default browser. |
| GH | Send search term or phrase to GitHub in default browser. |
| GO | Send search term or phrase Google in default browser. |
| GS | Send search term or phrase to Google Scholar in default browser. |
| GV | Send search term or phrase to Google Videos in default browser. |
| IPM | Read list of search terms and submit each term to PubMed in a separate browser tab. |
| MA | Send search term to all searchable websites in pymolshortcuts. |
| MB | Send search term to multiple sites that contain book content. |
| MC | Send search term to search ten core websites in pymolshortcuts: |
| MM | Send search term to search for manuscripts in pymolshortcuts. |
| PDB | Submit a search term to the Protein Data Bank. |
| PM | Send search term or phrase to PubMed. |
| PML | Submit a search term to the PyMOL Users Mail Service. |
| PW | Submit search of the PyMOL Wiki. |
| RG | Submit a search query of Research Gate. |
| SD | Submit a search term to Science Direct. |
| SF | Send search term to sourceforge. |
| SO | Submit a search term to Stackoverflow. |
| SP | Submit a search term to Springer Books |
| Shortcut | Short Description |
|---|---|
| BST | G2G3/U9U8 base step , PDB code 4PCO. |
| GGT | WT human gamma glutamyl transpeptidase at 1.67 Angstrom resolution as cartoon. PDB Code 4gdx. |
| GU | 10-mer dsRNA with 8 GU wobble base pairs. |
| LG | Nine sugar glycan in influenza N9 neuraminidase, PDB code 4dgr. |
| N9 | Influenza N9 neuraminidase at 1.55 Angstrom resolution, PDB code 4dgr. |
| NA | Hydrated sodium cation bound in major groove of a 16-mer RNA of Watson-Crick base pairs. |
| T4L | WT T4 lysozyme as ribbon diagram (1.08 Ang): 3FA0. |
| U8 | 16-mer dsRNA with 8 contiguous Us. U-helix RNA (1.37 Ang): 3nd3. |
| WC8 | 16-mer dsRNA, Watson-Crick helix RNA: 3nd4. |
| Shortcut | Short Description |
|---|---|
| LBST | G2G3/U9U8 base step , PDB code 4PCO. |
| LGGT | WT human gamma glutamyl transpeptidase as cartoon. PDB code 4gdx. |
| LGU | 10-mer dsRNA. |
| LLG | Nine sugar glycan in influenza N9 neuraminidase at 1.55 Angstrom resolution, PDB code 4dgr. |
| LN9 | Influenza N9 neuraminidase, PDB code 4dgr. |
| LNA | Hydrated sodium cation bound in major groove of RNA with 16 Watson-Crick base pairs. |
| LT4L | Display WT T4 lysozyme as ribbon diagram (resolution 1.08 Ang): 3FA0. |
| LU8 | 16-mer dsRNA with 8 contiguous Us. U-helix RNA (1.37 Ang): 3nd3. |
| LWC8 | 16-mer dsRNA, Watson-Crick helix RNA, 3nd4. |
| SC | Print to screen list of the shortcuts that are available in the script pymolshortcuts.py. |
| Shortcut | Short Description |
|---|---|
| sb | Show van der Waals clashes as red discs. |
| Shortcut | Short Description |
|---|---|
| iterm | Open iTerm2 window on MacOS. |
| term | Open a Terminal window on MacOS. |
| x11 | Open x11 terminal. |
| xquartz | Open new XQuartz terminal on MacOS. |
| Shortcut | Short Description |
|---|---|
| atom | Open the text editor Atom from within PyMOL. |
| bbedit | Open file with the text editor bbedit from within PyMOL. |
| code | Open file with Visual Studio Code from within PyMOL. |
| emacs | Open file with emacs from within PyMOL. |
| gedit | Open file with gedit from within PyMOL. |
| jedit | Open file with jedit from within PyMOL. |
| mate | Open textmate from within PyMOL. |
| npp | Open notepadpp from within PyMOL. |
| nv | Open neovim from within PyMOL. |
| oni | Open the editor Oni from within PyMOL. |
| pdbed | Open PDBEditor.jar from within PyMOL. |
| st3 | Open Sublime Text 3 from within PyMOL. |
| vim | Open vim from within PyMOL. |
| Shortcut | Short Description |
|---|---|
| gcal | Open Google Calendar. |
| gmail | Open gmail. |
| webmail | Open Web Mail in defualt browser. |
| Shortcut | Short Description |
|---|---|
| ACA | Open the American Crystallographic Association Annual Meeting webpage. |
| ALS | Open website of the Advanced Light Source. |
| APS | Open website of the Advanced Photon Source. |
| CHESS | Open the website of CHESS. |
| EMDB | Open the website of the Electron Microscopy Data Bank. |
| EP | EasyPyMOL github site. |
| IUCR | Open website of the IUCr Journals. |
| JM | Open the Jmol wiki. |
| LBSF | Open website of Laboratory of Biomolecular Structure and Function, the X-ray diffraction core facility at OUHSC. |
| MCL | Open website of Macromolecular Crystallography Laboratory at the University of Oklahoma. |
| MG | Open website of the OUHSC molecular graphics course. |
| MGW | Open Wikipedia webpage about molecular graphics. |
| NDB | Open website of the Nucleic Acid Database. |
| NSLSII | Open the website of the National Synchrotron Light Source II (NSLSII) at Brookhaven National Laboratory. |
| PPC | Open the website of the Protein Production Facility at the University of Oklahoma in Norman. |
| PS | Open the home page of the Protein Soceity. |
| RS | Open the homepage of the RNA Society. |
| SAXS | Open the webpage of SAXS links at OUHSC. |
| SSRLSMB | Open the webpage of SSRL Structural Molecular Biology. |
| SSURF | Open the webpage of the Society for Science at User Research Facilities (SSURF). |
| SciPy19 | Open the SciPy 2019 YouTube Channel. |
| biocat | Open the webpage of the BIOCAT biological SAXS beamline at the Advanced Photon Source. |
| chimeraWeb | Open the webste of UCSF Chimera. |
| sasbdb | Open the webpage of the Small Angle Scattering Biological Data Bank (SASBDB). |
| sbgrid | Open the webpage of the Structural Biology Grid (SBGRID) YouTube Channel. |
| ssrlbl42 | Open the webpage of SSRL Biological SAXS at BL 4-2. |
| weather | Open National Weather Service website for locale. |
| Shortcut | Short Description |
|---|---|
| word | Open word from within PyMOL. |
- easypymol
- pymolsnips
- orgpymolpysnips
- rstudiopymolpysnips
- taggedpymolpysnips
- jupyterlabpymolpysnips
- colabOpenSourcePyMOLpySnips
- colabPyMOLpySnips
- PyMOLwallhangings
Insert update history
| Version | Changes | Date |
|---|---|---|
| Version 0.2 | Added badges, funding, update table, and edited README.md | 2024 May 8 |
- NIH: R01 CA242845, R01 AI088011
- NIH: P30 CA225520 (PI: R. Mannel); P20GM103640 and P30GM145423 (PI: A. West)