-
Notifications
You must be signed in to change notification settings - Fork 1
RdRp
The viral RNA dependent RNA polymerase (RdRp) gene encodes for the RdRp protein responsible for copying the genome of an RNA virus. Almost all RNA viruses make an RdRp protein (Wikipedia article on RdRp). Exceptions include RNA retroviruses, where the replicase is a reverse transcriptase (RT), and satellite viruses such as Hepatitis Delta Virus which use cellular host polymerases or helper viruses for their replication.
For an extended discussion see: Edgar, 2023.

RdRp is not always called RdRp. For example, in nidoviruses the RdRp is encoded as part of ORF1ab and the protein is called Nsp9 or Nsp12, and in the lambda 3 phage it is simply called the lambda protein (see PDB 1MUK). This complicates database searches by gene name.
Sometimes, RdRp has a simple gene structure, it's coding sequence (CDS) begins with a START codon and ending at a STOP codon. In such cases, it is straightforward to identify RdRp as the nucleotide or translated amino acid sequence of this CDS (assuming you know the genetic code of the host). However, things are not always that simple. RdRp often occurs in a longer ORF which codes for multiple proteins; for example in SARS-CoV-2 RdRp function is found in Nsp12, one of several non-structural proteins coded in the long ORF1ab (21,555nt) which accounts for most of the genome (29,903nt). ORF1ab is translated into a long peptide (pp1ab), which is then split into mature proteins by a cleavage enzyme.
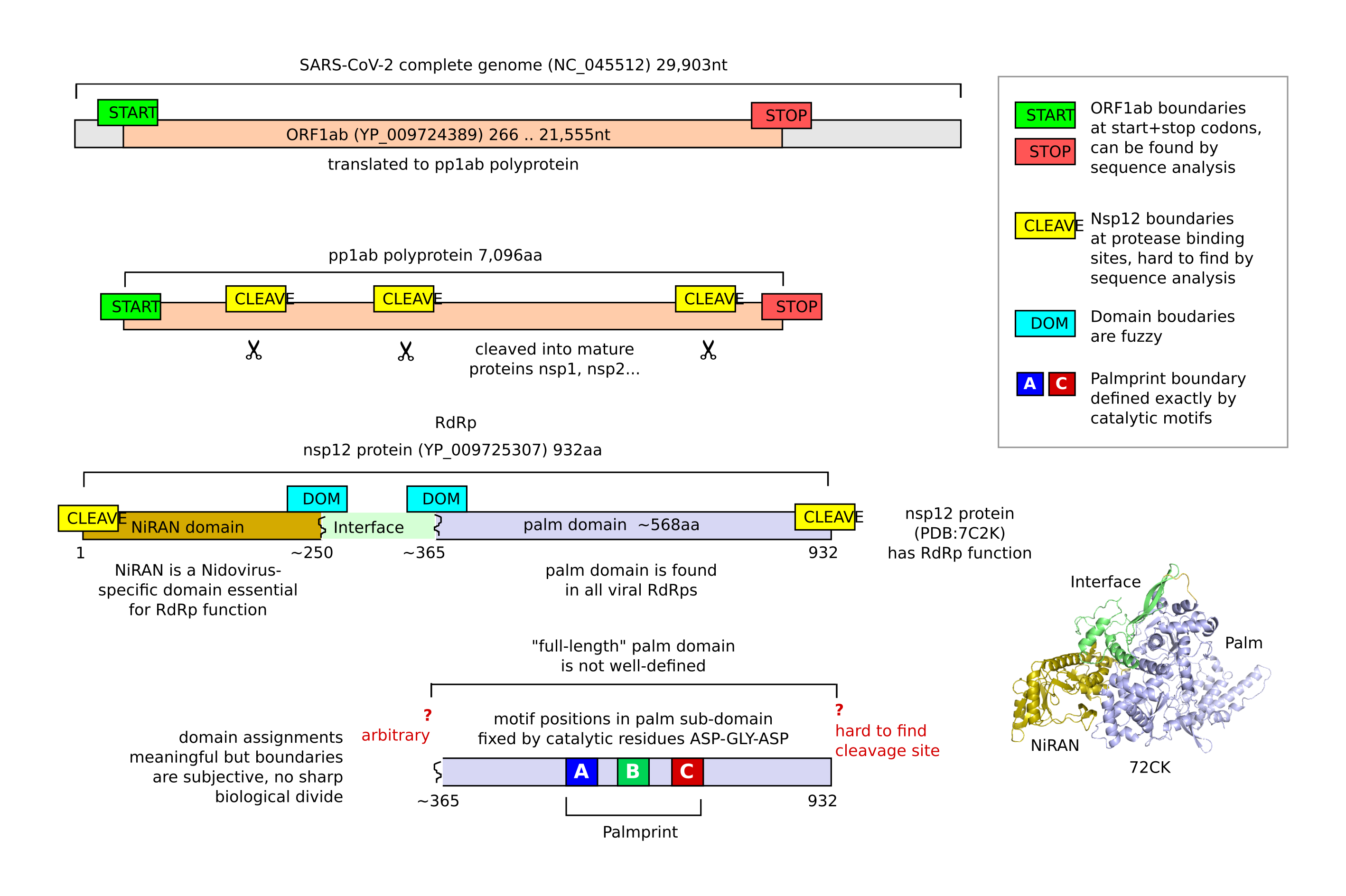
Unlike START and STOP codons, cleavage sites are not easily recognized by sequence analysis, and in fact are not known in many fully-sequenced genomes (https://pubmed.ncbi.nlm.nih.gov/27567259/. Therefore, the beginning and end of the RdRp sequence in a genome or metagenomic contig may be difficult to identify. In practice, this problem means that predicted RdRp sequences are often truncated or trimmed to shorter subsequences such as a partial domain.
The RdRp protein is usually constructed from two or more domains. The term domain generally means a segment which folds independently and may be found in combination with other domains in different proteins, but the definition is not precise and may be used in different ways in different contexts.
It can be clear that some residues are in a particular domain (for example, the [G/S]DD motif is the palm domain). However, there are no sharp boundaries where two adjacent residues are definitively in different domains. Thus, in contrast to CDS which has clear boundaries (start / stop codons), domain boundaries are inherently fuzzy.
The palm domain is universal among the known viral RdRps (Jia and Gong 2019, te Velthuis 2014). Other domains (named or not) can be found adjacent to the RdRp-palm domain; for example, nidoviruses such as SARS-CoV-2 have a NiRAN domain at the N terminal before the palm, and reoviruses have an unnamed N-terminal domain before the palm and a bracelet domain after the palm. Regions outside of the palm domain are generally not well understood, and it is often not known whether they are essential for RdRp function or perform some other function. This is a major confounder to analyzing RdRp since HMM models built from different groups of RdRp include adjacent domains which are incomparable outside of the group. For example SARS-CoV-2 RdRp (NiRAN + Palm) and Reovirus RdRp (Nterm + Palm + Bracelet) cannot be globally aligned to one another.

As the examples of nidoviruses (NiRAN+palm) and reoviruses (N-terminal+palm+bracelet) illustrate, the domain content of RdRp varies in different families, and except for the palm domain are often not well characterised. This adds to the difficulty of identifying the boundaries of the RdRp coding sequence in genomes which are far diverged from well-characterised viruses, and similarly in metagenomic contigs.
In at least one family (narnaviruses), RdRp can be encoded by more than one protein; it is assembled as a complex from preptides coded in multiple genome segments (https://pubmed.ncbi.nlm.nih.gov/34688782/).
Several families have permuted RdRp genes (Sabanadzovic2009, Ambrose2009, Ferrero2021, Gorbalenya2002) where the palm domain is permuted.