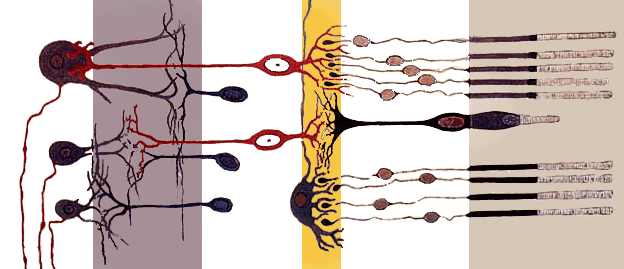
This is a prototype for an artificial neural environment using Erlang. It will be strongly biologically motivated.
The project is hosted at github
Some tennants
- be distributed/parallel/concurrent
- be biological
- see the forest and the trees
Forest and trees?
Or, maybe this?
Beware all ye who skip this quote; it's cool.
For one thing, Butler is not content to say that machines extend the organism, but asserts that they are really limbs and organs lying on the body without organs of a society, which men will appropriate according to their power and their wealth, and whose poverty deprives them as if they were mutilated organisms. For another, he is not content to say that organisms are machines, but asserts that they contain such an abundance of parts that they must be compared to very different parts of distinct machines, each relating to the others, engendered in combination with the others. [...] He shatters the vitalist argument by calling in question the specific or personal unity of the organism, and the mechanist argument even more decisively, by calling in question the structural unity of the machine.
- Deleuze and Guattari, Anti-Œdipus
Any erlang module (file) that has a start function should work as an example. As this gets more fleshed out I'll create an examples directory.
You'll need to compile any files you will use and any files the said files depend on (get it?). With Erlang you can do this at the command line like so
$ erlc file.erlwhich will generate the object file file.beam. However, since these files are currently meant to be used interactively in the Erlang VM you can just as easily compile files during your Erlang session like so
1> c(file).To use a given example you'll first need to understand the example (I'm serious). Then, you'll run the start function of the given example in the Erlang VM e.g. > example_module:start_function()..
With Erlang installed (in a Unixish system), we'll run a simple integrate and fire model.
Navigate to the code's directory and open the Erlang VM.
$ cd bane/examples
$ erlCompile and run the example then check for recieved messages.
Erlang R14B01 (erts-5.8.2) [source] [64-bit] [smp:2:2] [rq:2] [async-threads:0] [hipe] [kernel-poll:false]
Eshell V5.8.2 (abort with ^G)
1> c(simple).
{ok,simple}
2> simple:start().
{<0.38.0>,<0.39.0>}
3> flush().
Shell got {hi,{10,15,37}}
Shell got {hi,{10,15,38}}
Shell got {hi,{10,15,39}}
Shell got {hi,{10,15,40}}
Shell got {hi,{10,15,41}}
ok
4> q().
okThat's it. We flushed the messages we recieved (at the shell process) and noticed that we recieved one every second (in the time tuple {hour, min, sec}), which makes since, because our model has a single cell recieving input at 60Hz and with a threshold of 60 units (whatever our neurotransmitter quanta is).
- Wrap this in Autotools
- Figure out the correct way to install erlang libs (hmm)
- Extract common network libs from examples
- seperate examples from library from utilities?
- Workup an interface for I/O