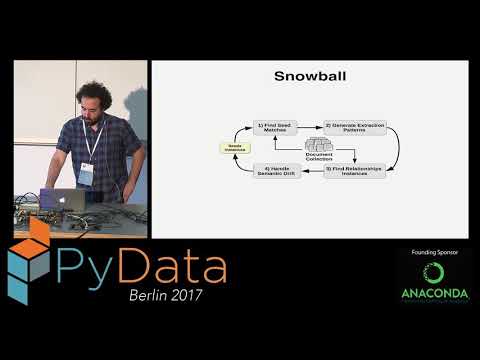
BREDS extracts relationships using a bootstrapping/semi-supervised approach, it relies on an initial set of seeds, i.e. pairs of named-entities representing relationship type to be extracted.
The algorithm expands the initial set of seeds using distributional semantics to generalize the relationship while limiting the semantic drift.
The input text needs to have the named-entities tagged, like show in the example bellow:
The tech company <ORG>Soundcloud</ORG> is based in <LOC>Berlin</LOC>, capital of Germany.
<ORG>Pfizer</ORG> says it has hired <ORG>Morgan Stanley</ORG> to conduct the review.
<ORG>Allianz</ORG>, based in <LOC>Munich</LOC>, said net income rose to EUR 1.32 billion.
<LOC>Switzerland</LOC> and <LOC>South Africa</LOC> are co-chairing the meeting.
<LOC>Ireland</LOC> beat <LOC>Italy</LOC> , then lost 43-31 to <LOC>France</LOC>.
<ORG>Pfizer</ORG>, based in <LOC>New York City</LOC> , employs about 90,000 workers.
<PER>Burton</PER> 's engine passed <ORG>NASCAR</ORG> inspection following the qualifying session.We need to give seeds to boostrap the extraction process, specifying the type of each named-entity and relationships examples that should also be present in the input text:
e1:ORG
e2:LOC
Lufthansa;Cologne
Nokia;Espoo
Google;Mountain View
DoubleClick;New York
SAP;WalldorfTo run a simple example, download the following files
- afp_apw_xin_embeddings.bin
- sentences_short.txt.bz2
- seeds_positive.txt
Install BREDS using pip
pip install breadsRun the following command:
breds --word2vec=afp_apw_xin_embeddings.bin --sentences=sentences_short.txt --positive_seeds=seeds_positive.txt --similarity=0.6 --confidence=0.6After the process is terminated an output file relationships.jsonl is generated containing the extracted relationships.
You can pretty print it's content to the terminal with: jq '.' < relationships.jsonl:
{
"entity_1": "Medtronic",
"entity_2": "Minneapolis",
"confidence": 0.9982486865148862,
"sentence": "<ORG>Medtronic</ORG> , based in <LOC>Minneapolis</LOC> , is the nation 's largest independent medical device maker . ",
"bef_words": "",
"bet_words": ", based in",
"aft_words": ", is",
"passive_voice": false
}
{
"entity_1": "DynCorp",
"entity_2": "Reston",
"confidence": 0.9982486865148862,
"sentence": "Because <ORG>DynCorp</ORG> , headquartered in <LOC>Reston</LOC> , <LOC>Va.</LOC> , gets 98 percent of its revenue from government work .",
"bef_words": "Because",
"bet_words": ", headquartered in",
"aft_words": ", Va.",
"passive_voice": false
}
{
"entity_1": "Handspring",
"entity_2": "Silicon Valley",
"confidence": 0.893486865148862,
"sentence": "There will be more firms like <ORG>Handspring</ORG> , a company based in <LOC>Silicon Valley</LOC> that looks as if it is about to become a force in handheld computers , despite its lack of machinery .",
"bef_words": "firms like",
"bet_words": ", a company based in",
"aft_words": "that looks",
"passive_voice": false
}BREDS has several parameters to tune the extraction process, in the example above it uses the default values, but these
can be set in the configuration file: parameters.cfg
max_tokens_away=6 # maximum number of tokens between the two entities
min_tokens_away=1 # minimum number of tokens between the two entities
context_window_size=2 # number of tokens to the left and right of each entity
alpha=0.2 # weight of the BEF context in the similarity function
beta=0.6 # weight of the BET context in the similarity function
gamma=0.2 # weight of the AFT context in the similarity function
wUpdt=0.5 # < 0.5 trusts new examples less on each iteration
number_iterations=4 # number of bootstrap iterations
wUnk=0.1 # weight given to unknown extracted relationship instances
wNeg=2 # weight given to extracted relationship instances
min_pattern_support=2 # minimum number of instances in a cluster to be considered a patternand passed with the argument --config=parameters.cfg.
The full command line parameters are:
-h, --help show this help message and exit
--config CONFIG file with bootstrapping configuration parameters
--word2vec WORD2VEC an embedding model based on word2vec, in the format of a .bin file
--sentences SENTENCES
a text file with a sentence per line, and with at least two entities per sentence
--positive_seeds POSITIVE_SEEDS
a text file with a seed per line, in the format, e.g.: 'Nokia;Espoo'
--negative_seeds NEGATIVE_SEEDS
a text file with a seed per line, in the format, e.g.: 'Microsoft;San Francisco'
--similarity SIMILARITY
the minimum similarity between tuples and patterns to be considered a match
--confidence CONFIDENCE
the minimum confidence score for a match to be considered a true positive
--number_iterations NUMBER_ITERATIONS
the number of iterations the runPlease, refer to the section References and Citations for details on the parameters.
In the first step, BREDS pre-processes the input file sentences.txt generating word vector representations of
relationships (i.e.: processed_tuples.pkl).
This is done so that then you can experiment with different seed examples without having to repeat the process of
generating word vectors representations. Just pass the argument --sentences=processed_tuples.pkl instead to skip
this generation step.
Semi-Supervised Bootstrapping of Relationship Extractors with Distributional Semantics, EMNLP'15
@inproceedings{batista-etal-2015-semi,
title = "Semi-Supervised Bootstrapping of Relationship Extractors with Distributional Semantics",
author = "Batista, David S. and Martins, Bruno and Silva, M{\'a}rio J.",
booktitle = "Proceedings of the 2015 Conference on Empirical Methods in Natural Language Processing",
month = sep,
year = "2015",
address = "Lisbon, Portugal",
publisher = "Association for Computational Linguistics",
url = "https://aclanthology.org/D15-1056",
doi = "10.18653/v1/D15-1056",
pages = "499--504",
}@incollection{phd-dsbatista2016
title = {Large-Scale Semantic Relationship Extraction for Information Discovery},
author = {Batista, David S.},
school = {Instituto Superior Técnico, Universidade de Lisboa},
year = {2016}
}Improvements, adding new features and bug fixes are welcome. If you wish to participate in the development of BREDS, please read the following guidelines.
- Preparing the development environment
- Code away!
- Continuous Integration
- Submit your changes by opening a pull request
Small fixes and additions can be submitted directly as pull requests, but larger changes should be discussed in an issue first. You can expect a reply within a few days, but please be patient if it takes a bit longer.
Make sure you have Python3.9 installed on your system
macOs
brew install python@3.9
python3.9 -m pip install --user --upgrade pip
python3.9 -m pip install virtualenvClone the repository and prepare the development environment:
git clone git@github.com:davidsbatista/BREDS.git
cd BREDS
python3.9 -m virtualenv venv # create a new virtual environment for development using python3.9
source venv/bin/activate # activate the virtual environment
pip install -r requirements_dev.txt # install the development requirements
pip install -e . # install BREDS in edit modeBREDS runs a continuous integration (CI) on all pull requests. This means that if you open a pull request (PR), a full test suite is run on your PR:
- The code is formatted using
blackandisort - Unused imports are auto-removed using
pycln - Linting is done using
pylingandflake8 - Type checking is done using
mypy - Tests are run using
pytest
Nevertheless, if you prefer to run the tests & formatting locally, it's possible too.
make allEvery PR should be accompanied by short description of the changes, including:
- Impact and motivation for the changes
- Any open issues that are closed by this PR
Give a ⭐️ if this project helped you!