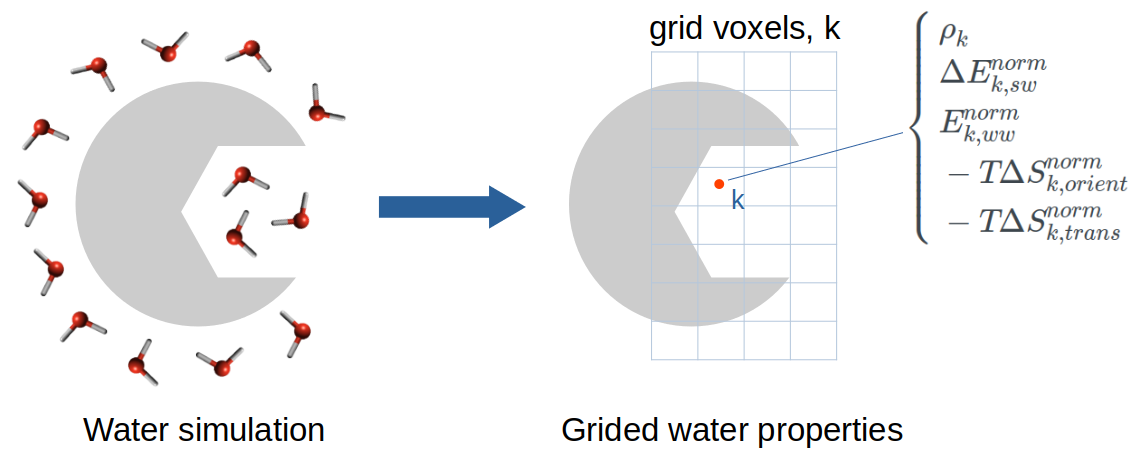
- BTK inhibitor: water replacement
- PDE10A inhibitor: as a solvation scoring function
- Identify the key pharmacohore feature
- Halogen bond interaction design
1. load the script into flare python gui: Flare | Python | Python Interpretor |Load .. (Figure 1. step 1, 2 and 3)
2. select the protein intereted (Figure 1. step 4).
3. pick a atom, single click button Run (Figure 1. step 5 and 6).
Figure 1. Five steps to use the script
- Yoshida, S.; Uehara, S.; Kondo, N.; Takahashi, Y.; Yamamoto, S.; Kameda, A.; Kawagoe, S.; Inoue, N.; Yamada, M.; Yoshimura, N.; et al. Peptide-to-Small Molecule: A Pharmacophore-Guided Small Molecule Lead Generation Strategy from High-Affinity Macrocyclic Peptides. 2022. https://doi.org/10.1021/acs.jmedchem.2c00919.
- Yang, Z.; Xiaoyun, L. U. The Role of Water Molecules in Drug Design. Prog. Pharm. Sci. 2022, 46 (1), 47–59. https://pps.cpu.edu.cn/article/id/ba95392b-7a5e-44d4-a2b5-a3084711d6f5
- Targowska-Duda, K. M.; Maj, M.; Drączkowski, P.; Budzyńska, B.; Boguszewska-Czubara, A.; Wróbel, T. M.; Laitinen, T.; Kaczmar, P.; Poso, A.; Kaczor, A. A. WaterMap Guided Structure‐based Virtual Screening for Acetylcholinesterase Inhibitors. ChemMedChem 2022, n/a (n/a). https://doi.org/10.1002/cmdc.202100721.
- Zsidó, B. Z.; Hetényi, C. The Role of Water in Ligand Binding. Curr. Opin. Struct. Biol. 2021, 67 (Figure 1), 1–8. https://doi.org/10.1016/j.sbi.2020.08.002.
- Lloyd, M. G.; Huckvale, R.; Cheung, K. J.; Rodrigues, M. J.; Collie, G. W.; Pierrat, O. A.; Gatti Iou, M.; Carter, M.; Davis, O. A.; McAndrew, P. C.; et al. Into Deep Water: Optimizing BCL6 Inhibitors by Growing into a Solvated Pocket. J. Med. Chem. 2021, 64 (23), 17079–17097. https://doi.org/10.1021/acs.jmedchem.1c00946.
- Hüfner-Wulsdorf, T.; Klebe, G. Mapping Water Thermodynamics on Drug Candidates via Molecular Building Blocks: A Strategy to Improve Ligand Design and Rationalize SAR. J. Med. Chem. 2021, 64 (8), 4662–4676. https://doi.org/10.1021/acs.jmedchem.0c02115.
- Andreev, S.; Pantsar, T.; Tesch, R.; Kahlke, N.; El-Gokha, A.; Ansideri, F.; Grätz, L.; Romasco, J.; Sita, G.; Geibel, C.; et al. Addressing a Trapped High-Energy Water: Design and Synthesis of Highly Potent Pyrimidoindole-Based Glycogen Synthase Kinase-3β Inhibitors. J. Med. Chem. 2021, acs.jmedchem.0c02146. https://doi.org/10.1021/acs.jmedchem.0c02146.
- Hüfner-Wulsdorf, T.; Klebe, G. Protein–Ligand Complex Solvation Thermodynamics: Development, Parameterization, and Testing of GIST-Based Solvent Functionals. J. Chem. Inf. Model. 2020, 60 (3), 1409–1423. https://doi.org/10.1021/acs.jcim.9b01109.
- Yoshidome, T.; Ikeguchi, M.; Ohta, M. Comprehensive 3D-RISM Analysis of the Hydration of Small Molecule Binding Sites in Ligand-Free Protein Structures. J. Comput. Chem. 2020, 41 (28), 2406–2419. https://doi.org/10.1002/jcc.26406.
- Hüfner-Wulsdorf, T.; Klebe, G. Advancing GIST-Based Solvent Functionals through Multiobjective Optimization of Solvent Enthalpy and Entropy Scoring Terms. J. Chem. Inf. Model. 2020, 60 (12), 6654–6665. https://doi.org/10.1021/acs.jcim.0c01133.
- Li, Y.; Gao, Y.; Holloway, M. K.; Wang, R. Prediction of the Favorable Hydration Sites in a Protein Binding Pocket and Its Application to Scoring Function Formulation. J. Chem. Inf. Model. 2020, 60 (9), 4359–4375. https://doi.org/10.1021/acs.jcim.9b00619.
- Viviani, L. G.; Piccirillo, E.; Ulrich, H.; Amaral, A. T. -d. T. D. Virtual Screening Approach for the Identification of Hydroxamic Acids as Novel Human Ecto-5′-Nucleotidase Inhibitors. J. Chem. Inf. Model. 2020, 60 (2), 621–630. https://doi.org/10.1021/acs.jcim.9b00884.
- Bancet, A.; Raingeval, C.; Lomberget, T.; Le Borgne, M.; Guichou, J.-F.; Krimm, I. Fragment Linking Strategies for Structure-Based Drug Design. J. Med. Chem. 2020, 63 (20), 11420–11435. https://doi.org/10.1021/acs.jmedchem.0c00242.
- Gerstenberger, B. S.; Ambler, C.; Arnold, E. P.; Banker, M.; Brown, M. F.; Clark, J. D.; Dermenci, A.; Dowty, M. E.; Fensome, A.; Fish, S.; et al. Discovery of Tyrosine Kinase 2 (TYK2) Inhibitor (PF-06826647) for the Treatment of Autoimmune Diseases. J. Med. Chem. 2020, 63 (22), 13561–13577. https://doi.org/10.1021/acs.jmedchem.0c00948.
- Wang, Y.; Fu, Q.; Zhou, Y.; Du, Y.; Huang, N. Replacement of Protein Binding-Site Waters Contributes to Favorable Halogen Bond Interactions. J. Chem. Inf. Model. 2019, 59 (7), 3136–3143. https://doi.org/10.1021/acs.jcim.9b00128.
- Schaller, D.; Pach, S.; Wolber, G. PyRod: Tracing Water Molecules in Molecular Dynamics Simulations. J. Chem. Inf. Model. 2019, No. May, acs.jcim.9b00281. https://doi.org/10.1021/acs.jcim.9b00281.
- Nittinger, E.; Gibbons, P.; Eigenbrot, C.; Davies, D. R.; Maurer, B.; Yu, C. L.; Kiefer, J. R.; Kuglstatter, A.; Murray, J.; Ortwine, D. F.; et al. Water Molecules in Protein–Ligand Interfaces. Evaluation of Software Tools and SAR Comparison. J. Comput. Aided. Mol. Des. 2019, 33 (3), 307–330. https://doi.org/10.1007/s10822-019-00187-y.
- Nguyen, C.; Yamazaki, T.; Kovalenko, A.; Case, D. A.; Gilson, M. K.; Kurtzman, T.; Luchko, T. A Molecular Reconstruction Approach to Site-Based 3D-RISM and Comparison to GIST Hydration Thermodynamic Maps in an Enzyme Active Site. PLoS One 2019, 14 (7), e0219473. https://doi.org/10.1371/journal.pone.0219473.
- Lu, J.; Hou, X.; Wang, C.; Zhang, Y. Incorporating Explicit Water Molecules and Ligand Conformation Stability in Machine-Learning Scoring Functions. J. Chem. Inf. Model. 2019, 59 (11), 4540–4549. https://doi.org/10.1021/acs.jcim.9b00645.
- Bucher, D.; Stouten, P.; Triballeau, N. Shedding Light on Important Waters for Drug Design: Simulations versus Grid-Based Methods. J. Chem. Inf. Model. 2018, 58 (3), 692–699. https://doi.org/10.1021/acs.jcim.7b00642.
- Nittinger, E.; Flachsenberg, F.; Bietz, S.; Lange, G.; Klein, R.; Rarey, M. Placement of Water Molecules in Protein Structures: From Large-Scale Evaluations to Single-Case Examples. J. Chem. Inf. Model. 2018, 58 (8), 1625–1637. https://doi.org/10.1021/acs.jcim.8b00271.
- Hamaguchi, H.; Amano, Y.; Moritomo, A.; Shirakami, S.; Nakajima, Y.; Nakai, K.; Nomura, N.; Ito, M.; Higashi, Y.; Inoue, T. Discovery and Structural Characterization of Peficitinib (ASP015K) as a Novel and Potent JAK Inhibitor. Bioorg. Med. Chem. 2018, 26 (18), 4971–4983. https://doi.org/10.1016/j.bmc.2018.08.005.
- Wang, Y.; Du, Y.; Huang, N. A Survey of the Role of Nitrile Groups in Protein–Ligand Interactions. Future Med. Chem. 2018, 10 (23), 2713–2728. https://doi.org/10.4155/fmc-2018-0252.
- Masters, M. R.; Mahmoud, A. H.; Yang, Y.; Lill, M. A. Efficient and Accurate Hydration Site Profiling for Enclosed Binding Sites. J. Chem. Inf. Model. 2018, 58 (11), 2183–2188. https://doi.org/10.1021/acs.jcim.8b00544.
- Ahmad, S.; Shaker, B.; Ahmad, F.; Raza, S.; Azam, S. S. Moleculer Dynamics Simulaiton Revealed Reciever Domain of Acinetobacter Baumannii BfmR Enzyme as the Hot Spot for Future Antibiotics Designing. J. Biomol. Struct. Dyn. 2018, 1–42. https://doi.org/10.1080/07391102.2018.1498805.
- Ahmad, S.; Raza, S.; Abro, A.; Liedl, K. R.; Azam, S. S. Toward Novel Inhibitors against KdsB: A Highly Specific and Selective Broad-Spectrum Bacterial Enzyme. J. Biomol. Struct. Dyn. 2018, 1–20. https://doi.org/10.1080/07391102.2018.1459318.
- Matter, H.; Güssregen, S. Characterizing Hydration Sites in Protein-Ligand Complexes towards the Design of Novel Ligands. Bioorg. Med. Chem. Lett. 2018, 28 (14), 2343–2352. https://doi.org/10.1016/j.bmcl.2018.05.061.
- Callegari, D.; Ranaghan, K. E.; Woods, C. J.; Minari, R.; Tiseo, M.; Mor, M.; Mulholland, A. J.; Lodola, A. L718Q Mutant EGFR Escapes Covalent Inhibition by Stabilizing a Non-Reactive Conformation of the Lung Cancer Drug Osimertinib. Chem. Sci. 2018, 9 (10), 2740–2749. https://doi.org/10.1039/C7SC04761D.
- Cappel, D.; Sherman, W.; Beuming, T. Calculating Water Thermodynamics in the Binding Site of Proteins – Applications of WaterMap to Drug Discovery. Curr. Top. Med. Chem. 2017, 17 (23), 2586–2598. https://doi.org/10.2174/1568026617666170414141452.
- Spyrakis, F.; Ahmed, M. H.; Bayden, A. S.; Cozzini, P.; Mozzarelli, A.; Kellogg, G. E. The Roles of Water in the Protein Matrix: A Largely Untapped Resource for Drug Discovery. J. Med. Chem. 2017, 60 (16), 6781–6828. https://doi.org/10.1021/acs.jmedchem.7b00057.
- Schauperl, M.; Czodrowski, P.; Fuchs, J. E.; Huber, R. G.; Waldner, B. J.; Podewitz, M.; Kramer, C.; Liedl, K. R. Binding Pose Flip Explained via Enthalpic and Entropic Contributions. J. Chem. Inf. Model. 2017, 57 (2), 345–354. https://doi.org/10.1021/acs.jcim.6b00483.
- Balius, T. E.; Fischer, M.; Stein, R. M.; Adler, T. B.; Nguyen, C. N.; Cruz, A.; Gilson, M. K.; Kurtzman, T.; Shoichet, B. K. Testing Inhomogeneous Solvation Theory in Structure-Based Ligand Discovery. Proc. Natl. Acad. Sci. 2017, 114 (33), E6839–E6846. https://doi.org/10.1073/pnas.1703287114.
- Güssregen, S.; Matter, H.; Hessler, G.; Lionta, E.; Heil, J.; Kast, S. M. Thermodynamic Characterization of Hydration Sites from Integral Equation-Derived Free Energy Densities: Application to Protein Binding Sites and Ligand Series. J. Chem. Inf. Model. 2017, 57 (7), 1652–1666. https://doi.org/10.1021/acs.jcim.6b00765.
- Czodrowski, P.; Mallinger, A.; Wienke, D.; Esdar, C.; Pöschke, O.; Busch, M.; Rohdich, F.; Eccles, S. A.; Ortiz-Ruiz, M.-J.; Schneider, R.; et al. Structure-Based Optimization of Potent, Selective, and Orally Bioavailable CDK8 Inhibitors Discovered by High-Throughput Screening. J. Med. Chem. 2016, 59 (20), 9337–9349. https://doi.org/10.1021/acs.jmedchem.6b00597.
- Bodnarchuk, M. S. Water, Water, Everywhere... It’s Time to Stop and Think. Drug Discov. Today 2016, 21 (7), 1139–1146. https://doi.org/10.1016/j.drudis.2016.05.009.
- Myrianthopoulos, V.; Gaboriaud-Kolar, N.; Tallant, C.; Hall, M.-L.; Grigoriou, S.; Brownlee, P. M.; Fedorov, O.; Rogers, C.; Heidenreich, D.; Wanior, M.; et al. Discovery and Optimization of a Selective Ligand for the Switch/Sucrose Nonfermenting-Related Bromodomains of Polybromo Protein-1 by the Use of Virtual Screening and Hydration Analysis. J. Med. Chem. 2016, 59 (19), 8787–8803. https://doi.org/10.1021/acs.jmedchem.6b00355.
- Ramsey, S.; Nguyen, C.; Salomon-Ferrer, R.; Walker, R. C.; Gilson, M. K.; Kurtzman, T. Solvation Thermodynamic Mapping of Molecular Surfaces in AmberTools: GIST. J. Comput. Chem. 2016, 37 (21), 2029–2037. https://doi.org/10.1002/jcc.24417.
- Calabrò, G.; Woods, C. J.; Powlesland, F.; Mey, A. S. J. S.; Mulholland, A. J.; Michel, J. Elucidation of Nonadditive Effects in Protein–Ligand Binding Energies: Thrombin as a Case Study. J. Phys. Chem. B 2016, 120 (24), 5340–5350. https://doi.org/10.1021/acs.jpcb.6b03296.
- Zoidis, G.; Giannakopoulou, E.; Stevaert, A.; Frakolaki, E.; Myrianthopoulos, V.; Fytas, G.; Mavromara, P.; Mikros, E.; Bartenschlager, R.; Vassilaki, N.; et al. Novel Indole–Flutimide Heterocycles with Activity against Influenza PA Endonuclease and Hepatitis C Virus. Medchemcomm 2016, 7 (3), 447–456. https://doi.org/10.1039/C5MD00439J.
- Murphy, R. B.; Repasky, M. P.; Greenwood, J. R.; Tubert-Brohman, I.; Jerome, S.; Annabhimoju, R.; Boyles, N. A.; Schmitz, C. D.; Abel, R.; Farid, R.; et al. WScore: A Flexible and Accurate Treatment of Explicit Water Molecules in Ligand-Receptor Docking. J. Med. Chem. 2016, 59 (9), 4364–4384. https://doi.org/10.1021/acs.jmedchem.6b00131.
- Uehara, S.; Tanaka, S. AutoDock-GIST: Incorporating Thermodynamics of Active-Site Water into Scoring Function for Accurate Protein-Ligand Docking. Molecules 2016, 21 (11), 1604. https://doi.org/10.3390/molecules21111604.
- Horbert, R.; Pinchuk, B.; Johannes, E.; Schlosser, J.; Schmidt, D.; Cappel, D.; Totzke, F.; Schächtele, C.; Peifer, C. Optimization of Potent Dfg-in Inhibitors of Platelet Derived Growth Factor Receptorβ (PDGF-Rβ) Guided by Water Thermodynamics. J. Med. Chem. 2015, 58 (1), 170–182. https://doi.org/10.1021/jm500373x.
- Bayden, A. S.; Moustakas, D. T.; Joseph-McCarthy, D.; Lamb, M. L. Evaluating Free Energies of Binding and Conservation of Crystallographic Waters Using SZMAP. J. Chem. Inf. Model. 2015, 55 (8), 1552–1565. https://doi.org/10.1021/ci500746d.
- Nguyen, C. N.; Cruz, A.; Gilson, M. K.; Kurtzman, T. Thermodynamics of Water in an Enzyme Active Site: Grid-Based Hydration Analysis of Coagulation Factor Xa. chemrxiv 2014, 10 (7), 2769–2780. https://doi.org/10.1021/ct401110x.
- Czodrowski, P.; Hölzemann, G.; Barnickel, G.; Greiner, H.; Musil, D. Selection of Fragments for Kinase Inhibitor Design: Decoration Is Key. J. Med. Chem. 2015, 58 (1), 457–465. https://doi.org/10.1021/jm501597j.
- Smith, C. R.; Dougan, D. R.; Komandla, M.; Kanouni, T.; Knight, B.; Lawson, J. D.; Sabat, M.; Taylor, E. R.; Vu, P.; Wyrick, C. Fragment-Based Discovery of a Small Molecule Inhibitor of Bruton’s Tyrosine Kinase. J. Med. Chem. 2015, 58 (14), 5437–5444. https://doi.org/10.1021/acs.jmedchem.5b00734.
- Bortolato, A.; Tehan, B. G.; Bodnarchuk, M. S.; Essex, J. W.; Mason, J. S. Water Network Perturbation in Ligand Binding: Adenosine A 2A Antagonists as a Case Study. J. Chem. Inf. Model. 2013, 53 (7), 1700–1713. https://doi.org/10.1021/ci4001458.
- Breiten, B.; Lockett, M. R.; Sherman, W.; Fujita, S.; Al-Sayah, M.; Lange, H.; Bowers, C. M.; Heroux, A.; Krilov, G.; Whitesides, G. M. Water Networks Contribute to Enthalpy/Entropy Compensation in Protein–Ligand Binding. J. Am. Chem. Soc. 2013, 135 (41), 15579–15584. https://doi.org/10.1021/ja4075776.
- Woods, C. J.; Malaisree, M.; Long, B.; McIntosh-Smith, S.; Mulholland, A. J. Computational Assay of H7n9 Influenza Neuraminidase Reveals R292k Mutation Reduces Drug Binding Affinity. Sci. Rep. 2013, 3, 7–12. https://doi.org/10.1038/srep03561.
- Kohlmann, A.; Zhu, X.; Dalgarno, D. Application of MM-GB/SA and WaterMap to SRC Kinase Inhibitor Potency Prediction. ACS Med. Chem. Lett. 2012, 3 (2), 94–99. https://doi.org/10.1021/ml200222u.
- Beuming, T.; Che, Y.; Abel, R.; Kim, B.; Shanmugasundaram, V.; Sherman, W. Thermodynamic Analysis of Water Molecules at the Surface of Proteins and Applications to Binding Site Prediction and Characterization. Proteins Struct. Funct. Bioinforma. 2012, 80 (3), 871–883. https://doi.org/10.1002/prot.23244.
- Trujillo, J. I.; Kiefer, J. R.; Huang, W.; Day, J. E.; Moon, J.; Jerome, G. M.; Bono, C. P.; Kornmeier, C. M.; Williams, M. L.; Kuhn, C.; et al. Investigation of the Binding Pocket of Human Hematopoietic Prostaglandin (PG) D2 Synthase (HH-PGDS): A Tale of Two Waters. Bioorg. Med. Chem. Lett. 2012, 22 (11), 3795–3799. https://doi.org/10.1016/j.bmcl.2012.04.004.
- Mason, J. S.; Bortolato, A.; Congreve, M.; Marshall, F. H. New Insights from Structural Biology into the Druggability of G Protein-Coupled Receptors. Trends Pharmacol. Sci. 2012, 33 (5), 249–260. https://doi.org/10.1016/j.tips.2012.02.005.
- Kung, P.-P.; Sinnema, P.-J.; Richardson, P.; Hickey, M. J.; Gajiwala, K. S.; Wang, F.; Huang, B.; McClellan, G.; Wang, J.; Maegley, K.; et al. Design Strategies to Target Crystallographic Waters Applied to the Hsp90 Molecular Chaperone. Bioorg. Med. Chem. Lett. 2011, 21 (12), 3557–3562. https://doi.org/10.1016/j.bmcl.2011.04.130.
- Nguyen, C.; Gilson, M. K.; Young, T. Structure and Thermodynamics of Molecular Hydration via Grid Inhomogeneous Solvation Theory. ChemRxiv 2011.
- Robinson, D. D.; Sherman, W.; Farid, R. Understanding Kinase Selectivity Through Energetic Analysis of Binding Site Waters. ChemMedChem 2010, 5 (4), 618–627. https://doi.org/10.1002/cmdc.200900501.
- Michel, J.; Tirado-Rives, J.; Jorgensen, W. L. Energetics of Displacing Water Molecules from Protein Binding Sites: Consequences for Ligand Optimization. J. Am. Chem. Soc. 2009, 131 (42), 15403–15411. https://doi.org/10.1021/ja906058w.
- Abel, R.; Young, T.; Farid, R.; Berne, B. J.; Friesner, R. A. Role of the Active-Site Solvent in the Thermodynamics of Factor Xa Ligand Binding. J. Am. Chem. Soc. 2008, 130 (9), 2817–2831. https://doi.org/10.1021/ja0771033.
- Young, T.; Abel, R.; Kim, B.; Berne, B. J.; Friesner, R. A. Motifs for Molecular Recognition Exploiting Hydrophobic Enclosure in Protein–Ligand Binding. Proc. Natl. Acad. Sci. 2007, 104 (3), 808–813. https://doi.org/10.1073/pnas.0610202104.
- Chen, J. M.; Xu, S. L.; Wawrzak, Z.; Basarab, G. S.; Jordan, D. B. Structure-Based Design of Potent Inhibitors of Scytalone Dehydratase: Displacement of a Water Molecule from the Active Site. Biochemistry 1998, 37 (51), 17735–17744. https://doi.org/10.1021/bi981848r.