-
Notifications
You must be signed in to change notification settings - Fork 5k
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Jupyter terminal using different sys.path from Jupyter notebook (running on same server, same conda env) #3311
Comments
|
You've installed a kernelspec at some point which points to the other environment. Run |
|
Update: |
|
Have you installed Jupyter in the env you're talking about? |
|
With the conda env activated, This issue was ultimately resolved by installing the package Thanks for your help! |
|
I had the same issue, and nb_conda fixed it for me. Essentially, as whittredge explained, Jupyter was looking into a path different than my virtual environment. After installing nb_conda, I was able to start Jupyter notebook and then select the specific kernel I wanted it to access (in my case, the specific virtual env I had created). |
|
Hi, The issues I encounter are only in the notebook and not in the terminal. The terminal shows correct paths. |
|
I was able to solve the problem by installing jupyter notebook within my new environment. |
|
delete |
|
Thanks @swissrapperswil. I have both Canopy and Anaconda, and due to an update, the default switched to Canopy. For this reason, the conda packages, while running smoothly using bash would be unrecognizable in jupyter notebooks. Simply installing the jupyter notebook in the default (canopy) environment solved it. |
Actually, such a problem comes when you have conda environment and you install jupyter via pip instead of conda. It happened to me, I had jupyter running but couldn't import libraries. |
|
Changing the executable path of jupyter (in the |
This saved my life! Thank you so much! |
|
I have the same issue but, at the same time, kind of different.
Any suggestions? |
|
I am having a similar issue except the terminal gives I find it so puzzling that the notebook and the terminal launched from within jupyter point to different environments! So strange! |
|
The notebook information reflects the kernel's environment, while the terminal information reflects that of the notebook server. I suspect your kernel configuration is launching a python relative to your base environment. Run This is probably similar to #3311 (comment) (now that I see that response above), although it appears that that solution still uses an absolute path - which should be fine as well. If you take the relative approach I suggest above, the resolution is probably more a function of your |
|
For people using the strategy of installing jupyter notebook within the environment and then running it, it may be worth to check which jupyter you are activating by default. Run first |
|
Would like to second @engmubarak48 ; this problem was fixed for me when I changed the environment.yaml file I was using to generate the environment to include |
|
Had similar issue, only I was using pipenv as virtual environment. |
Thanks, I encountered the same problem, and resolved by installing `nb_conda''. :) |
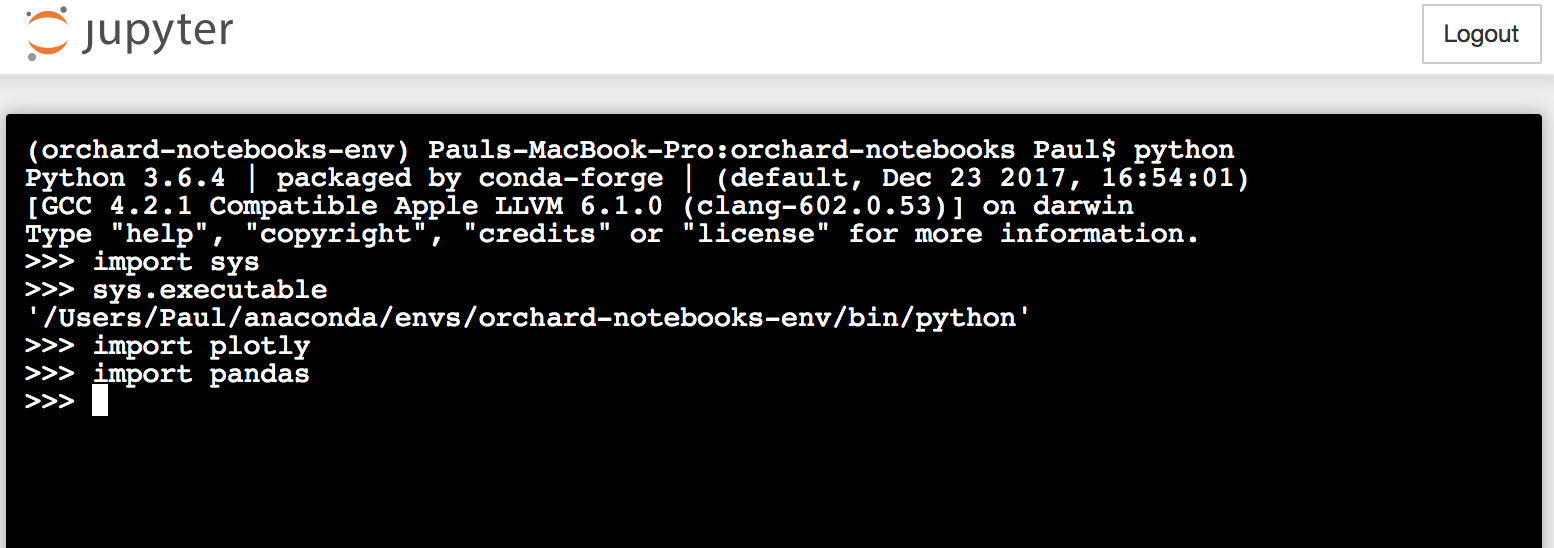



I was having a common package import issue that was mostly resolved by several similar issues (#2359, #397, #2898), but now I have a new issue that I don't think has been mentioned here. I am using conda on macOS 10.12.
If activate a conda env with
source activate $envname, then runpythonfrom my local terminal, I can easily import the packages that I have installed in my envname.yml file. From this conda env I can start ajupyter notebookand run a Jupyter terminal. In the Jupyter terminal,sys.executablereturns the same result as in my local terminal, and I can import all the same packages. HOWEVER, if I start a new Jupyter notebook from the same launcher, running side-by-side the functioning Jupyter terminal on the same server,sys.executablereturns a DIFFERENT path (one that I thought I had deleted). Of course, this means I cannot import the package I hope to use in the notebook. Not only that, but the list ofsys.pathlocations in the Jupyter terminal versus the Jupyter notebook are completely non-overlapping---see screenshots below.It seems from the above screenshots that the Jupyter terminal is choosing to run python from a different conda env than the Jupyter notebook. To solve this problem I removed the wrong env that the notebook was using as sys.path with
conda env remove. Now, there are no kernel errors when I start the Jupyter launcher or a Jupyter terminal, but starting a notebook returns a kernel error, as if it is expecting to work only with the path listed in the removed conda env.The text was updated successfully, but these errors were encountered: