Top-1% solution to the Cassava Leaf Disease Classification Kaggle competition on plant image classification.
Cassava is one of the key food crops grown in Africa. Plant diseases are major sources of poor yields. To diagnose plant diseases, farmers require the help of agricultural experts to visually inspect the plants, which is labor-intensive and costly. Deep learning helps to automate this process.
This project works with a dataset of 21,367 cassava images. The pictures are taken by farmers on mobile phones and labeled as healthy or having one of the four common leaf disease types. Main data-related challenges are poor image quality, inconsistent background conditions and label noise.
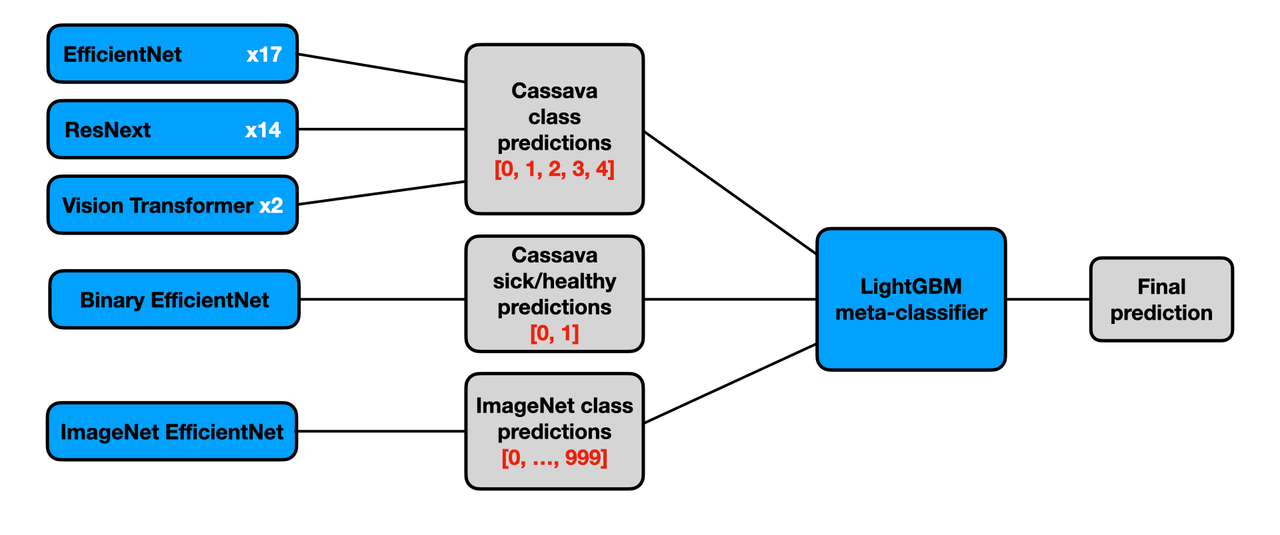
We develop a stacking ensemble with CNNs and Vision Transformers implemented in PyTorch. Our solution reaches the test accuracy of 91.06% and places 14th out of 3,900 competing teams. The diagram below overviews the ensemble. The detailed summary of the solution is provided this writeup.
The project has the following structure:
codes/:.pyscripts with training, inference and image processing functionsnotebooks/:.ipynbnotebooks for data eploration, training CNN/ViT models and ensemblingdata/: input data (images are not included due to size constraints and can be downloaded here)output/: model weights, configurations and diagrams exported from notebookspretraining/: weights and configurations of models pretrained on external datasets
To execute codes, you can create a virtual Conda environment from the environment.yml file:
conda env create --name cassava --file environment.yml
conda activate cassava
The solution can be reproduced in the following steps:
- Downloading competition data and adding it into the
data/folder. - Running training notebooks
pytorch-modelto obtain base models weights. - Running the ensembling notebook
lightgbm-stackingto get final predictions.
All pytorch-model notebooks have the same structure and differ in model/data parameters. Different versions are included to ensure reproducibility. If you only wish to get familiar with our solution, it is enough to inspect one of the modeling notebooks and go through codes/ to understand the training process. The stacking ensemble reproducing our submission is also provided in this Kaggle notebook.
The notebooks are designed to run on Google Colab. More details are provided in the documentation within the notebooks.