use SVM and PCA_ResNet50 to classify HSI
===== update:
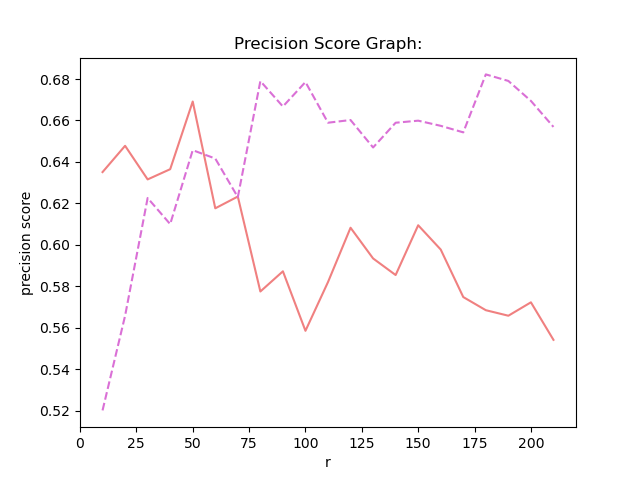
Besides using NMF method , then i use GNMF method , and i compare the result between NMF and GNMF.
result:
-
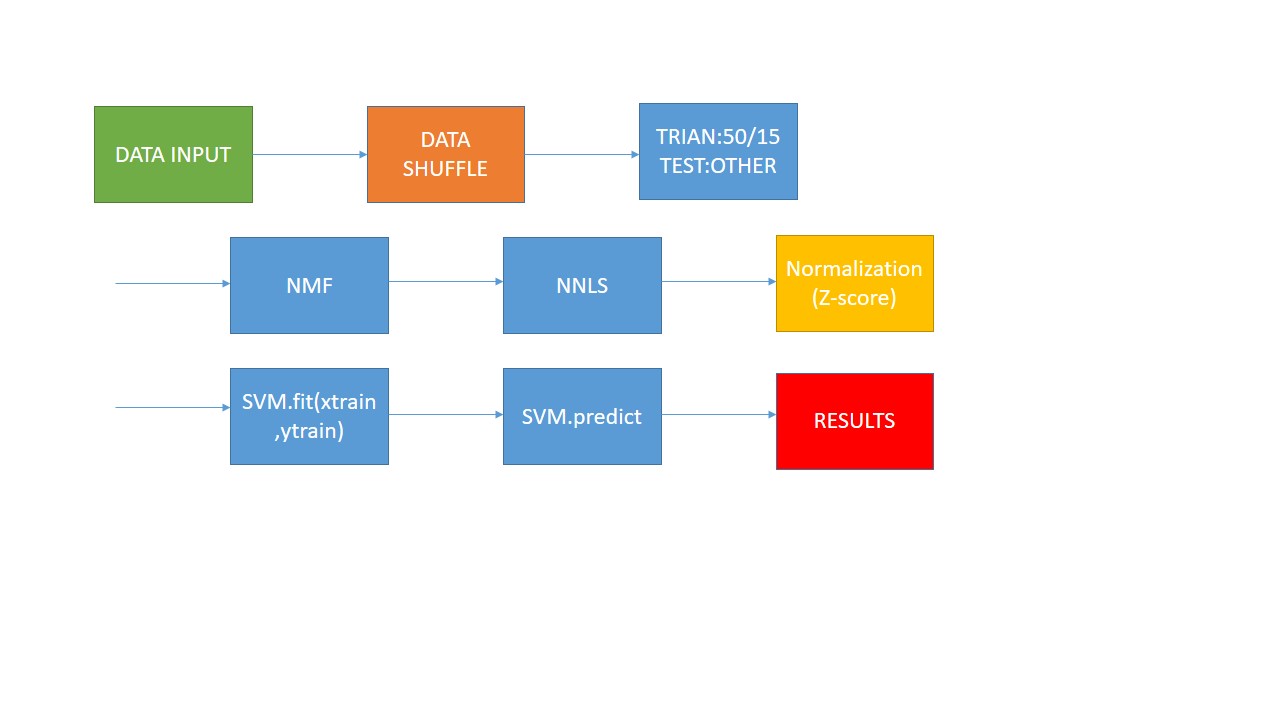
stage1: use SVM to classify HSI(Hyperspectral Image). opertion : Firstly , transfer 3D Indian pines data to 2D data , and the origial groundtruth data is 2D data , I transfer it to 1D data. Then, according the quantity of GT(groundTruth) , I choose 15 vectors from the first three least and choose 50 vectors from the other vectors. Treat these data as training data.the left data is test data. And I use the NMF as the method of demension reduction . Before NMF , I normalize the data using Z-Score. Then i use NNLS on the W matrix AFTER NMF and get H matrix. Finally i use SVM to fit and predict the label. This is the NNLS.
-
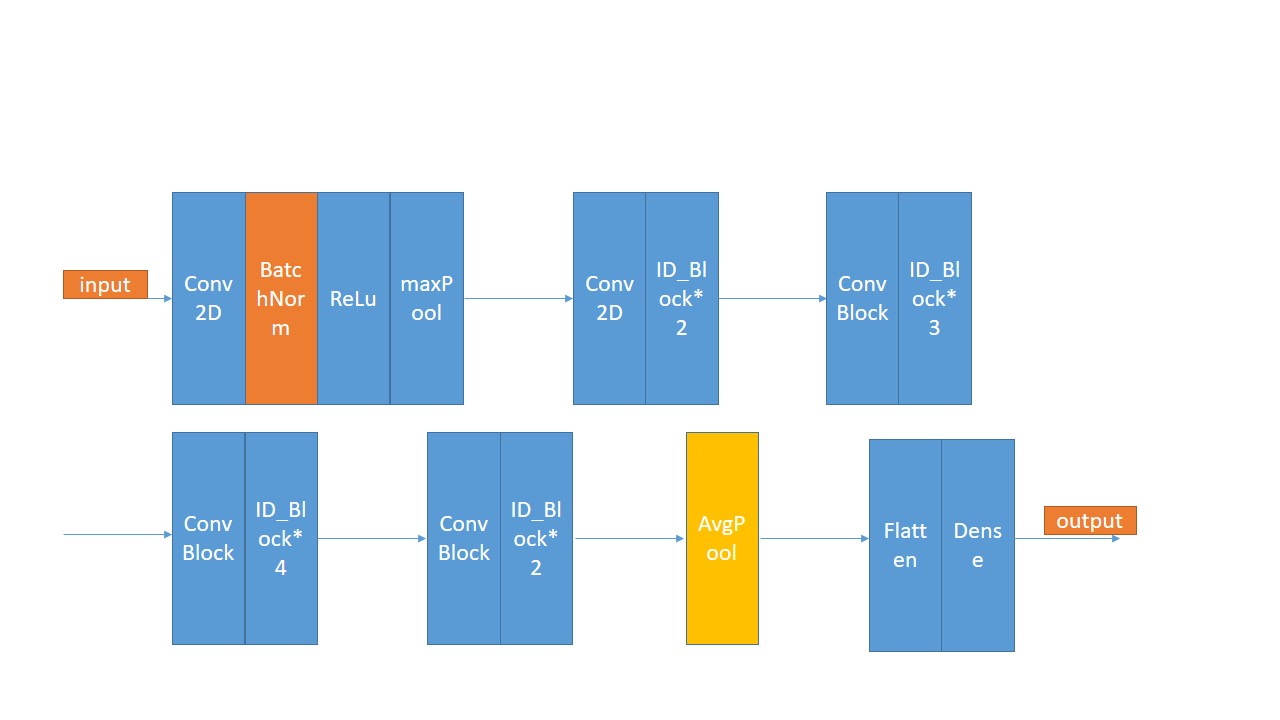
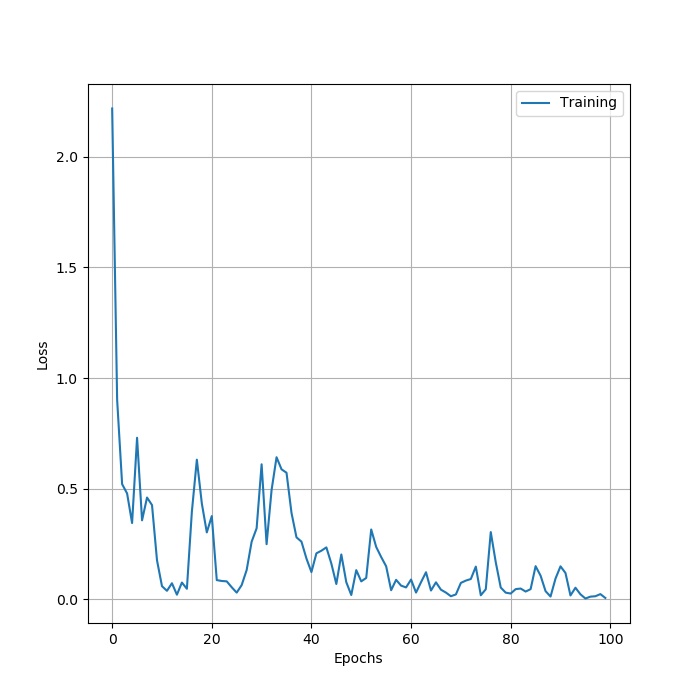
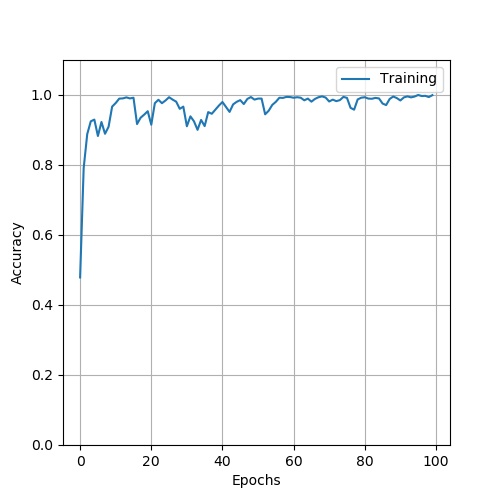
stage2: use PCA and ResNet50 to classify HSI. Firstly,choose 20% of the Indian pines data as the training data , the other is the test data. Use PCA as the method of demension reduction , after I put the data into ResNet50 , finally get the resut. Here is the structure of ResNet50
. The result: loss:
. accuary:
Reference: PCA ResNet50_on_Keras HybridSN
-
how to use it: run :
pip install -r requirements.txtfirst to install the packages. if you want to use SVM , you can run
python train_demo.py.If you want to use PCA+ResNet50 , you can run
python PCA_ResNet50.py -
contact: if you have problems , you can pick up an issue and communicate with me , here is my wechat number:Yingbin192