Imagine the situation: You’ve written some wonderful Python code which produces a beautiful graph as an output. You save that graph, naturally enough, as graph.png. You run the code a couple of times, each time making minor modifications. You come back to it the next week/month/year. Do you know how you created that graph? What input data? What version of your code? If you’re anything like me then the answer will often, frustratingly, be “no”. Of course, you then waste lots of time trying to work out how you created it, or even give up and never use it in that journal paper that will win you a Nobel Prize…
This talk will introduce ReciPy (from recipe and python), a Python module that will save you from this situation! (Although it can’t guarantee that your resulting paper will win a Nobel Prize!) With the addition of a single line of code to the top of your Python files, ReciPy will log each run of your code to a database, keeping track of the input files, output files and the version of your code, and then let you query this database to find out how you actually did create graph.png.
The easiest way to install is by simply running
pip install recipy
Alternatively, you can clone this repository and run:
python setup.py install
If you want to install the dependencies manually (they should be installed automatically if you're following the instructions above) then run:
pip install -r requirements.txt
You can upgrade from a previous release by running:
pip install -U recipy
To find out what has changed since the last release, see the changelog
Note: Previous (unreleased) versions of recipy required MongoDB to be installed and set up manually. This is no longer required, as a pure Python database (TinyDB) is used instead. Also, the GUI is now integrated fully into recipy and does not require installing separately.
Simply add the following line to the top of your Python script:
import recipyNote that this must be the very top line of your script, before you import anything else.
Then just run your script as usual, and all of the data will be logged into the TinyDB database (don't worry, the database is automatically created if needed). You can then use the recipy script to quickly query the database to find out what run of your code produced what output file. So, for example, if you run some code like this:
import recipy
import numpy
arr = numpy.arange(10)
arr = arr + 500
numpy.save('test.npy', arr)(Note the addition of import recipy at the beginning of script - but there are no other changes from a standard script)
Alternatively, run an unmodified script with python -m recipy SCRIPT [ARGS ...] to enable recipy logging. This invokes recipy's module entry point, which takes care of import recipy for you, before running your script.
it will produce an output called test.npy. To find out the details of the run which created this file you can search using
recipy search test.npy
and it will display information like the following:
Created by robin on 2015-05-25 19:00:15.631000
Ran /Users/robin/code/recipy/example_script.py using /usr/local/opt/python/bin/python2.7
Git: commit 91a245e5ea82f33ae58380629b6586883cca3ac4, in repo /Users/robin/code/recipy, with origin git@github.com:recipy/recipy.git
Environment: Darwin-14.3.0-x86_64-i386-64bit, python 2.7.9 (default, Feb 10 2015, 03:28:08)
Inputs:
Outputs:
/Users/robin/code/recipy/test.npy
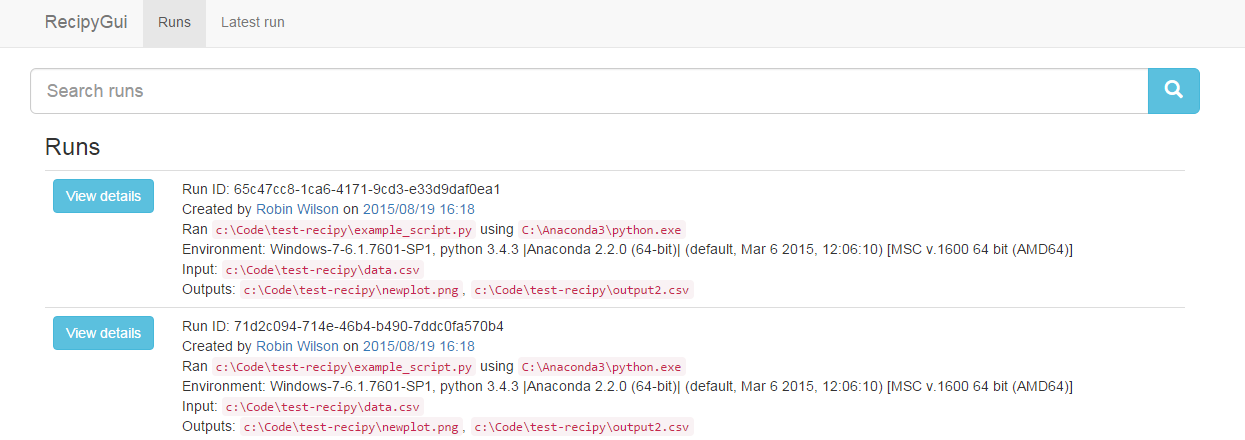
An alternative way to view this is to use the GUI. Just run recipy gui and a browser window will open with an interface that you can use to search all of your recipy 'runs':
If you want to log inputs and outputs of files read or written with built-in open, you need to do a little more work. Either use recipy.open (only requires import recipy at the top of your script), or add from recipy import open and just use open.
This workaround is required, because many libraries use built-in open internally, and you only want to record the files you explicitly opened yourself.
If you use Python 2, you can pass an encoding parameter to recipy.open. In this case codecs is used to open the file with proper encoding.
Once you've got some runs in your database, you can 'annotate' these runs with any notes that you want to keep about them. This can be particularly useful for recording which runs worked well, or particular problems you ran into. This can be done from the 'details' page in the GUI, or by running
recipy annotate
which will open an editor to allow you to write notes that will be attached to the run. These will then be viewable via the command-line and the GUI when searching for runs.
There are other features in the command-line interface too: recipy --help to see the other options. You can view diffs, see all runs that created a file with a given name, search based on ids, show the latest entry and more:
recipy - a frictionless provenance tool for Python
Usage:
recipy search [options] <outputfile>
recipy latest [options]
recipy gui [options]
recipy annotate [<idvalue>]
recipy (-h | --help)
recipy --version
Options:
-h --help Show this screen
--version Show version
-a --all Show all results (otherwise just latest result given)
-f --fuzzy Use fuzzy searching on filename
-r --regex Use regex searching on filename
-i --id Search based on (a fragment of) the run ID
-v --verbose Be verbose
-d --diff Show diff
-j --json Show output as JSON
--no-browser Do not open browser window
--debug Turn on debugging mode
Recipy stores all of its configuration and the database itself in ~/.recipy. Recipy's main configuration file is inside this folder, called recipyrc. The configuration file format is very simple, and is based on Windows INI files - and having a configuration file is completely optional: the defaults will work fine with no configuration file.
An example configuration is:
[ignored metadata]
diff
[general]
debug
This simply instructs recipy not to save git diff information when it records metadata about a run, and also to print debug messages (which can be really useful if you're trying to work out why certain functions aren't patched). At the moment, the only possible options are:
[general]debug- print debug messageseditor = vi- Configure the default text editor that will be used when recipy needs you to type in a message. Use notepad if on Windows, for examplequiet- don't print any messagesport- specify port to use for the GUI
[data]file_diff_outputs- store diff between the old output and new output file, if the output file exists before the script is executed
[database]path = /path/to/file.json- set the path to the database file
[ignored metadata]diff- don't store the output ofgit diffin the metadata for a recipy rungit- don't store anything relating to git (origin, commit, repo etc) in the metadata for a recipy runinput_hashes- don't compute and store SHA-1 hashes of input filesoutput_hashes- don't compute and store SHA-1 hashes of output files
[ignored inputs]- List any module here (eg.
numpy) to instruct recipy not to record inputs from this module, orallto ignore inputs from all modules
- List any module here (eg.
[ignored outputs]- List any module here (eg.
numpy) to instruct recipy not to record outputs from this module, orallto ignore outputs from all modules
- List any module here (eg.
By default all metadata is stored (ie. no metadata is ignored) and debug messages are not shown. A .recipyrc file in the current directory takes precedence over the ~/.recipy/recipyrc file, allowing per-project configurations to be easily handled.
Note: No default configuration file is provided with recipy, so if you wish to configure anything you will need to create a properly-formatted file yourself.
When you import recipy it adds a number of classes to sys.meta_path. These are then used by Python as part of the importing procedure for modules. The classes that we add are classes derived from PatchImporter, often using the easier interface provided by PatchSimple, which allow us to wrap functions that do input/output in a function that calls recipy first to log the information.
Generally, most of the complexity is hidden away in PatchImporter and PatchSimple (plus utils.py), so the actual code to wrap a module, such as numpy is fairly simple:
# Inherit from PatchSimple
class PatchNumpy(PatchSimple):
# Specify the full name of the module
modulename = 'numpy'
# List functions that are involved in input/output
# these can be anything that can go after "modulename."
# so they could be something like "pyplot.savefig" for example
input_functions = ['genfromtxt', 'loadtxt', 'load', 'fromfile']
output_functions = ['save', 'savez', 'savez_compressed', 'savetxt']
# Define the functions that will be used to wrap the input/output
# functions.
# In this case we are calling the log_input function to log it to the DB
# and we are giving it the 0th argument from the function (because all of
# the functions above take the filename as the 0th argument), and telling
# it that it came from numpy.
input_wrapper = create_wrapper(log_input, 0, 'numpy')
output_wrapper = create_wrapper(log_output, 0, 'numpy')A class like this must be implemented for each module whose input/output needs logging. At the moment the following input and output functions are patched:
This table lists the modules recipy has patches for, and the input and output functions that are patched.
| Module | Input functions | Output functions |
|---|---|---|
pandas |
read_csv, read_table, read_excel, read_hdf, read_pickle, read_stata, read_msgpack |
DataFrame.to_csv, DataFrame.to_excel, DataFrame.to_hdf, DataFrame.to_msgpack, DataFrame.to_stata, DataFrame.to_pickle, Panel.to_excel, Panel.to_hdf, Panel.to_msgpack, Panel.to_pickle, Series.to_csv, Series.to_hdf, Series.to_msgpack, Series.to_pickle |
matplotlib.pyplot |
savefig |
|
numpy |
genfromtxt, loadtxt, fromfile |
save, savez, savez_compressed, savetxt |
lxml.etree |
parse, iterparse |
|
bs4 |
BeautifulSoup |
|
gdal |
Open |
Driver.Create, Driver.CreateCopy |
sklearn |
datasets.load_svmlight_file |
datasets.dump_svmlight_file |
nibabel |
nifti1.Nifti1Image.from_filename, nifti2.Nifti2Image.from_filename, freesurfer.mghformat.MGHImage.from_filename, spm99analyze.Spm99AnalyzeImage.from_filename, minc1.Minc1Image.from_filename, minc2.Minc2Image.from_filename, analyze.AnalyzeImage.from_filename, parrec.PARRECImage.from_filename, spm2analyze.Spm2AnalyzeImage.from_filename |
nifti1.Nifti1Image.to_filename, nifti2.Nifti2Image.to_filename, freesurfer.mghformat.MGHImage.to_filename, spm99analyze.Spm99AnalyzeImage.to_filename, minc1.Minc1Image.to_filename, minc2.Minc2Image.to_filename, analyze.AnalyzeImage.to_filename, parrec.PARRECImage.to_filename, spm2analyze.Spm2AnalyzeImage.to_filename |
However, the code example above shows how easy it is to write a class to wrap a new module - so please feel free to submit a Pull Request to make recipy work with your favourite scientific modules!
recipy's test framework is in integration_test. The test framework has been designed to run under both Python 2.7+ and Python 3+. For more information see recipy test framework.
The test framework is run on the following platforms: