-
Notifications
You must be signed in to change notification settings - Fork 653
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
FIX-#3686: optimize __getitem__ flow of loc/iloc
#3694
Changes from 7 commits
4d30205
44e747e
afd87af
dbe3af7
a2f3d7f
40885a3
bf042ca
813037b
e9ba2d5
File filter
Filter by extension
Conversations
Jump to
Diff view
Diff view
There are no files selected for viewing
| Original file line number | Diff line number | Diff line change |
|---|---|---|
|
|
@@ -32,9 +32,10 @@ | |
| import numpy as np | ||
| import pandas | ||
| from pandas.api.types import is_list_like, is_bool | ||
| from pandas.core.dtypes.common import is_integer | ||
| from pandas.core.dtypes.common import is_integer, is_bool_dtype, is_integer_dtype | ||
| from pandas.core.indexing import IndexingError | ||
| from modin.error_message import ErrorMessage | ||
| from itertools import compress | ||
|
|
||
| from .dataframe import DataFrame | ||
| from .series import Series | ||
|
|
@@ -126,9 +127,34 @@ def is_boolean_array(x): | |
| bool | ||
| True if argument is an array of bool, False otherwise. | ||
| """ | ||
| if isinstance(x, (np.ndarray, Series, pandas.Series, pandas.Index)): | ||
| return is_bool_dtype(x.dtype) | ||
| elif isinstance(x, (DataFrame, pandas.DataFrame)): | ||
| return all(map(is_bool_dtype, x.dtypes)) | ||
| return is_list_like(x) and all(map(is_bool, x)) | ||
|
|
||
|
|
||
| def is_integer_array(x): | ||
| """ | ||
| Check that argument is an array of integers. | ||
|
|
||
| Parameters | ||
| ---------- | ||
| x : object | ||
| Object to check. | ||
|
|
||
| Returns | ||
| ------- | ||
| bool | ||
| True if argument is an array of integers, False otherwise. | ||
| """ | ||
| if isinstance(x, (np.ndarray, Series, pandas.Series, pandas.Index)): | ||
| return is_integer_dtype(x.dtype) | ||
| elif isinstance(x, (DataFrame, pandas.DataFrame)): | ||
| return all(map(is_integer_dtype, x.dtypes)) | ||
| return is_list_like(x) and all(map(is_integer, x)) | ||
|
|
||
|
|
||
| def is_integer_slice(x): | ||
| """ | ||
| Check that argument is an array of int. | ||
|
|
@@ -175,6 +201,32 @@ def is_range_like(obj): | |
| ) | ||
|
|
||
|
|
||
| def boolean_mask_to_numeric(indexer): | ||
| """ | ||
| Convert boolean mask to numeric indices. | ||
|
|
||
| Parameters | ||
| ---------- | ||
| indexer : list-like of booleans | ||
|
|
||
| Returns | ||
| ------- | ||
| np.ndarray of ints | ||
| Numerical positions of ``True`` elements in the passed `indexer`. | ||
| """ | ||
| if isinstance(indexer, (np.ndarray, Series, pandas.Series)): | ||
| return np.where(indexer)[0] | ||
| else: | ||
| # It's faster to build the resulting numpy array from the reduced amount of data via | ||
| # `compress` iterator than convert non-numpy-like `indexer` to numpy and apply `np.where`. | ||
| return np.fromiter( | ||
| # `itertools.compress` masks `data` with the `selectors` mask, | ||
| # works about ~10% faster than a pure list comprehension | ||
| compress(data=range(len(indexer)), selectors=indexer), | ||
dchigarev marked this conversation as resolved.
Show resolved
Hide resolved
|
||
| dtype=np.int64, | ||
| ) | ||
|
|
||
|
|
||
| _ILOC_INT_ONLY_ERROR = """ | ||
| Location based indexing can only have [integer, integer slice (START point is | ||
| INCLUDED, END point is EXCLUDED), listlike of integers, boolean array] types. | ||
|
|
@@ -505,6 +557,43 @@ def _parse_row_and_column_locators(self, tup): | |
| col_loc = col_loc(self.df) if callable(col_loc) else col_loc | ||
| return row_loc, col_loc, _compute_ndim(row_loc, col_loc) | ||
|
|
||
| # HACK: This method bypasses regular ``loc/iloc.__getitem__`` flow in order to ensure better | ||
| # performance in the case of boolean masking. The only purpose of this method is to compensate | ||
| # for a lack of backend's indexing API, there is no Query Compiler method allowing masking | ||
| # along both axis when any of the indexers is a boolean. That's why rows and columns masking | ||
| # phases are separate in this case. | ||
| # TODO: Remove this method and handle this case naturally via ``loc/iloc.__getitem__`` flow | ||
YarShev marked this conversation as resolved.
Show resolved
Hide resolved
|
||
| # when QC API would support both-axis masking with boolean indexers. | ||
| def _handle_boolean_masking(self, row_loc, col_loc): | ||
| """ | ||
| Retrieve dataset according to the boolean mask for rows and an indexer for columns. | ||
|
|
||
| In comparison with the regular ``loc/iloc.__getitem__`` flow this method efficiently | ||
| masks rows with a Modin Series boolean mask without materializing it (if the selected | ||
| execution implements such masking). | ||
|
|
||
| Parameters | ||
| ---------- | ||
| row_loc : modin.pandas.Series of bool dtype | ||
| Boolean mask to index rows with. | ||
| col_loc : object | ||
| An indexer along column axis. | ||
|
|
||
| Returns | ||
| ------- | ||
| modin.pandas.DataFrame or modin.pandas.Series | ||
| Located dataset. | ||
| """ | ||
| ErrorMessage.catch_bugs_and_request_email( | ||
| failure_condition=not isinstance(row_loc, Series), | ||
| extra_log=f"Only ``modin.pandas.Series`` boolean masks are acceptable, got: {type(row_loc)}", | ||
| ) | ||
| masked_df = self.df.__constructor__( | ||
| query_compiler=self.qc.getitem_array(row_loc._query_compiler) | ||
| ) | ||
| # Passing `slice(None)` as a row indexer since we've just applied it | ||
| return type(self)(masked_df)[(slice(None), col_loc)] | ||
|
|
||
|
|
||
| class _LocIndexer(_LocationIndexerBase): | ||
| """ | ||
|
|
@@ -539,32 +628,22 @@ def __getitem__(self, key): | |
| row_loc, col_loc, ndim = self._parse_row_and_column_locators(key) | ||
| self.row_scalar = is_scalar(row_loc) | ||
| self.col_scalar = is_scalar(col_loc) | ||
| if isinstance(row_loc, slice) and row_loc == slice(None): | ||
| # If we're only slicing columns, handle the case with `__getitem__` | ||
| if not isinstance(col_loc, slice): | ||
| # Boolean indexers can just be sliced into the columns object and | ||
| # then passed to `__getitem__` | ||
| if is_boolean_array(col_loc): | ||
| col_loc = self.df.columns[col_loc] | ||
| return self.df.__getitem__(col_loc) | ||
| else: | ||
| result_slice = self.df.columns.slice_locs(col_loc.start, col_loc.stop) | ||
| return self.df.iloc[:, slice(*result_slice)] | ||
|
Comment on lines
-542
to
-552
There was a problem hiding this comment. Choose a reason for hiding this commentThe reason will be displayed to describe this comment to others. Learn more. This is no longer needed, now the natural flow of This comment is no longer relevant: Reproducerimport modin.pandas as pd
import numpy as np
import timeit
shapes = [
# Wide frame
(5000, 5000),
# Long frame
(10_000_000, 10),
]
NRUNS = 100
for shape in shapes:
NROWS, NCOLS = shape
df = pd.DataFrame({f"col{i}": np.arange(NROWS) for i in range(NCOLS)})
print(f"Shape: {shape}")
print(f"__getitem__: {timeit.timeit(lambda: repr(df[['col1']]), number=NRUNS)}")
print(f".loc: {timeit.timeit(lambda: repr(df.loc[:, ['col1']]), number=NRUNS)}")
print(f".loc: {timeit.timeit(lambda: repr(df.loc[:, ['col1']]), number=NRUNS)}")
print(f"__getitem__: {timeit.timeit(lambda: repr(df[['col1']]), number=NRUNS)}")Also, check the results of ASV runs with the |
||
|
|
||
| if isinstance(row_loc, Series) and is_boolean_array(row_loc): | ||
| return self._handle_boolean_masking(row_loc, col_loc) | ||
|
|
||
| row_lookup, col_lookup = self._compute_lookup(row_loc, col_loc) | ||
| if any(i == -1 for i in row_lookup) or any(i == -1 for i in col_lookup): | ||
| raise KeyError( | ||
| "Passing list-likes to .loc or [] with any missing labels is no longer " | ||
| "supported, see https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#deprecate-loc-reindex-listlike" | ||
| ) | ||
|
Comment on lines
-555
to
-559
There was a problem hiding this comment. Choose a reason for hiding this commentThe reason will be displayed to describe this comment to others. Learn more. this check was moved to the |
||
| result = super(_LocIndexer, self).__getitem__(row_lookup, col_lookup, ndim) | ||
| if isinstance(result, Series): | ||
| result._parent = self.df | ||
| result._parent_axis = 0 | ||
| col_loc_as_list = [col_loc] if self.col_scalar else col_loc | ||
| row_loc_as_list = [row_loc] if self.row_scalar else row_loc | ||
| # Pandas drops the levels that are in the `loc`, so we have to as well. | ||
| if hasattr(result, "index") and isinstance(result.index, pandas.MultiIndex): | ||
| if ( | ||
| isinstance(result, (Series, DataFrame)) | ||
| and result._query_compiler.has_multiindex() | ||
|
There was a problem hiding this comment. Choose a reason for hiding this commentThe reason will be displayed to describe this comment to others. Learn more. checking for MultiIndex with the special method helps to avoid delayed computations triggering in the case of OmniSci backend There was a problem hiding this comment. Choose a reason for hiding this commentThe reason will be displayed to describe this comment to others. Learn more. this would be good as a code comment, not as a PR comment There was a problem hiding this comment. Choose a reason for hiding this commentThe reason will be displayed to describe this comment to others. Learn more. I'm not sure about this certain case. IMO this comment is excessive and would just bloat the if-statement. Any How this if-statement looks with the comment added# Option 1:
if (
isinstance(result, (Series, DataFrame))
# Checking for MultiIndex with the special method helps
# to avoid delayed computations triggering
and result._query_compiler.has_multiindex()
):
# Option 2:
if (
# Checking for MultiIndex with the special method helps
# to avoid delayed computations triggering
isinstance(result, (Series, DataFrame))
and result._query_compiler.has_multiindex()
):
# Option 3:
# Checking for MultiIndex with the special method helps
# to avoid delayed computations triggering
if (
isinstance(result, (Series, DataFrame))
and result._query_compiler.has_multiindex()
): |
||
| ): | ||
| if ( | ||
| isinstance(result, Series) | ||
| and not isinstance(col_loc_as_list, slice) | ||
|
|
@@ -583,7 +662,7 @@ def __getitem__(self, key): | |
| if ( | ||
| hasattr(result, "columns") | ||
| and not isinstance(col_loc_as_list, slice) | ||
| and isinstance(result.columns, pandas.MultiIndex) | ||
| and result._query_compiler.has_multiindex(axis=1) | ||
| and all( | ||
| col_loc_as_list[i] in result.columns.levels[i] | ||
| for i in range(len(col_loc_as_list)) | ||
|
|
@@ -689,49 +768,82 @@ def _compute_lookup(self, row_loc, col_loc): | |
|
|
||
| Returns | ||
| ------- | ||
| row_lookup : numpy.ndarray | ||
| row_lookup : slice(None) if full axis grab, pandas.RangeIndex if repetition is detected, numpy.ndarray otherwise | ||
| List of index labels. | ||
| col_lookup : numpy.ndarray | ||
| col_lookup : slice(None) if full axis grab, pandas.RangeIndex if repetition is detected, numpy.ndarray otherwise | ||
| List of columns labels. | ||
| """ | ||
| row_loc = [row_loc] if is_scalar(row_loc) else row_loc | ||
| col_loc = [col_loc] if is_scalar(col_loc) else col_loc | ||
| if is_list_like(row_loc) and len(row_loc) == 1: | ||
| if ( | ||
| isinstance(self.qc.index.values[0], np.datetime64) | ||
| and type(row_loc[0]) != np.datetime64 | ||
| ): | ||
| row_loc = [pandas.to_datetime(row_loc[0])] | ||
|
|
||
| if isinstance(row_loc, slice): | ||
| row_lookup = self.qc.index.get_indexer_for( | ||
| self.qc.index.to_series().loc[row_loc] | ||
| ) | ||
| elif self.qc.has_multiindex(): | ||
| if isinstance(row_loc, pandas.MultiIndex): | ||
| row_lookup = self.qc.index.get_indexer_for(row_loc) | ||
| else: | ||
| row_lookup = self.qc.index.get_locs(row_loc) | ||
| elif is_boolean_array(row_loc): | ||
| # If passed in a list of booleans, we return the index of the true values | ||
| row_lookup = [i for i, row_val in enumerate(row_loc) if row_val] | ||
| else: | ||
| row_lookup = self.qc.index.get_indexer_for(row_loc) | ||
| if isinstance(col_loc, slice): | ||
| col_lookup = self.qc.columns.get_indexer_for( | ||
| self.qc.columns.to_series().loc[col_loc] | ||
| ) | ||
| elif self.qc.has_multiindex(axis=1): | ||
| if isinstance(col_loc, pandas.MultiIndex): | ||
| col_lookup = self.qc.columns.get_indexer_for(col_loc) | ||
| Notes | ||
| ----- | ||
| Usage of `slice(None)` as a resulting lookup is a hack to pass information about | ||
| full-axis grab without computing actual indices that triggers lazy computations. | ||
| Ideally, this API should get rid of using slices as indexers and either use a | ||
| common ``Indexer`` object or range and ``np.ndarray`` only. | ||
| """ | ||
| lookups = [] | ||
| for axis, axis_loc in enumerate((row_loc, col_loc)): | ||
| if is_scalar(axis_loc): | ||
| axis_loc = np.array([axis_loc]) | ||
| if isinstance(axis_loc, slice) or is_range_like(axis_loc): | ||
| if isinstance(axis_loc, slice) and axis_loc == slice(None): | ||
| axis_lookup = axis_loc | ||
| else: | ||
| axis_labels = self.qc.get_axis(axis) | ||
| # `slice_indexer` returns a fully-defined numeric slice for a non-fully-defined labels-based slice | ||
| axis_lookup = axis_labels.slice_indexer( | ||
| axis_loc.start, axis_loc.stop, axis_loc.step | ||
| ) | ||
| # Converting negative indices to its actual positions: | ||
dchigarev marked this conversation as resolved.
Show resolved
Hide resolved
|
||
| axis_lookup = pandas.RangeIndex( | ||
| start=( | ||
| axis_lookup.start | ||
| if axis_lookup.start >= 0 | ||
| else axis_lookup.start + len(axis_labels) | ||
| ), | ||
vnlitvinov marked this conversation as resolved.
Show resolved
Hide resolved
|
||
| stop=( | ||
| axis_lookup.stop | ||
| if axis_lookup.stop >= 0 | ||
| else axis_lookup.stop + len(axis_labels) | ||
| ), | ||
| step=axis_lookup.step, | ||
| ) | ||
| elif self.qc.has_multiindex(axis): | ||
| # `Index.get_locs` raises an IndexError by itself if missing labels were provided, | ||
| # we don't have to do missing-check for the received `axis_lookup`. | ||
| if isinstance(axis_loc, pandas.MultiIndex): | ||
| axis_lookup = self.qc.get_axis(axis).get_indexer_for(axis_loc) | ||
| else: | ||
| axis_lookup = self.qc.get_axis(axis).get_locs(axis_loc) | ||
| elif is_boolean_array(axis_loc): | ||
| axis_lookup = boolean_mask_to_numeric(axis_loc) | ||
| else: | ||
| col_lookup = self.qc.columns.get_locs(col_loc) | ||
| elif is_boolean_array(col_loc): | ||
| # If passed in a list of booleans, we return the index of the true values | ||
| col_lookup = [i for i, col_val in enumerate(col_loc) if col_val] | ||
| else: | ||
| col_lookup = self.qc.columns.get_indexer_for(col_loc) | ||
| return row_lookup, col_lookup | ||
| axis_labels = self.qc.get_axis(axis) | ||
| if is_list_like(axis_loc) and not isinstance( | ||
| axis_loc, (np.ndarray, pandas.Index) | ||
| ): | ||
| # `Index.get_indexer_for` works much faster with numpy arrays than with python lists, | ||
| # so although we lose some time here on converting to numpy, `Index.get_indexer_for` | ||
| # speedup covers the loss that we gain here. | ||
| axis_loc = np.array(axis_loc, dtype=axis_labels.dtype) | ||
| axis_lookup = axis_labels.get_indexer_for(axis_loc) | ||
| # `Index.get_indexer_for` sets -1 value for missing labels, we have to verify whether | ||
| # there are any -1 in the received indexer to raise a KeyError here. | ||
| missing_mask = axis_lookup < 0 | ||
| if missing_mask.any(): | ||
| missing_labels = ( | ||
| # Converting `axis_loc` to maskable `np.array` to not fail | ||
| # on masking non-maskable list-like | ||
| np.array(axis_loc)[missing_mask] | ||
|
There was a problem hiding this comment. Choose a reason for hiding this commentThe reason will be displayed to describe this comment to others. Learn more.
|
||
| if is_list_like(axis_loc) | ||
| else axis_loc | ||
vnlitvinov marked this conversation as resolved.
Show resolved
Hide resolved
|
||
| ) | ||
| raise KeyError(f"Missing labels were provided: {missing_labels}") | ||
dchigarev marked this conversation as resolved.
Show resolved
Hide resolved
|
||
|
|
||
| if isinstance(axis_lookup, pandas.Index) and not is_range_like(axis_lookup): | ||
| axis_lookup = axis_lookup.values | ||
vnlitvinov marked this conversation as resolved.
Show resolved
Hide resolved
|
||
|
|
||
| lookups.append(axis_lookup) | ||
| return lookups | ||
|
|
||
|
|
||
| class _iLocIndexer(_LocationIndexerBase): | ||
|
|
@@ -770,6 +882,9 @@ def __getitem__(self, key): | |
| self._check_dtypes(row_loc) | ||
| self._check_dtypes(col_loc) | ||
|
|
||
| if isinstance(row_loc, Series) and is_boolean_array(row_loc): | ||
| return self._handle_boolean_masking(row_loc, col_loc) | ||
|
|
||
| row_lookup, col_lookup = self._compute_lookup(row_loc, col_loc) | ||
| result = super(_iLocIndexer, self).__getitem__(row_lookup, col_lookup, ndim) | ||
| if isinstance(result, Series): | ||
|
|
@@ -833,25 +948,43 @@ def _compute_lookup(self, row_loc, col_loc): | |
| Ideally, this API should get rid of using slices as indexers and either use a | ||
| common ``Indexer`` object or range and ``np.ndarray`` only. | ||
| """ | ||
| row_loc = [row_loc] if is_scalar(row_loc) else row_loc | ||
| col_loc = [col_loc] if is_scalar(col_loc) else col_loc | ||
| lookups = [] | ||
| for axis, axis_loc in enumerate((row_loc, col_loc)): | ||
| if isinstance(axis_loc, slice) and axis_loc.step is None: | ||
| if is_scalar(axis_loc): | ||
| axis_loc = np.array([axis_loc]) | ||
| if isinstance(axis_loc, slice): | ||
| axis_lookup = ( | ||
| axis_loc | ||
| if axis_loc == slice(None) | ||
| else pandas.RangeIndex( | ||
| *axis_loc.indices(len(self.qc.get_axis(axis))) | ||
| ) | ||
| ) | ||
| else: | ||
| axis_lookup = ( | ||
| pandas.RangeIndex(len(self.qc.get_axis(axis))) | ||
| .to_series() | ||
| .iloc[axis_loc] | ||
| .index | ||
| elif is_range_like(axis_loc): | ||
| axis_lookup = pandas.RangeIndex( | ||
| axis_loc.start, axis_loc.stop, axis_loc.step | ||
| ) | ||
| elif is_boolean_array(axis_loc): | ||
| axis_lookup = boolean_mask_to_numeric(axis_loc) | ||
| else: | ||
| if isinstance(axis_loc, pandas.Index): | ||
| axis_loc = axis_loc.values | ||
| elif is_list_like(axis_loc) and not isinstance(axis_loc, np.ndarray): | ||
| # `Index.__getitem__` works much faster with numpy arrays than with python lists, | ||
| # so although we lose some time here on converting to numpy, `Index.__getitem__` | ||
| # speedup covers the loss that we gain here. | ||
| axis_loc = np.array(axis_loc, dtype=np.int64) | ||
|
Comment on lines
+975
to
+978
There was a problem hiding this comment. Choose a reason for hiding this commentThe reason will be displayed to describe this comment to others. Learn more. Reproducerimport numpy as np
import timeit
from matplotlib import pyplot as plt
import pandas
N = 1_000_000
NRUNS = 5
index = pandas.RangeIndex(N)
def get_positions(index, percentage):
n_elements = int(len(index) * percentage)
indexer = list(range(n_elements))
return list(np.random.choice(indexer, n_elements, replace=False))
fns = {
"RangeIndex.__getitem__(python_list)": lambda index, indexer: index[indexer],
"RangeIndex.__getitem__(np.array(python_list))": lambda index, indexer: index[
np.array(indexer, dtype=np.int64)
],
}
X = np.arange(0, 101, 10)
results = {key: [] for key in fns.keys()}
for x in X:
print(f"{x}%")
python_list = get_positions(index=index, percentage=x / 100)
print(f"\tDEBUG: list length is {len(python_list)}")
for name, fn in fns.items():
print(f"\tRunning '{name}' fn...")
time = timeit.timeit(lambda: fn(index, python_list), number=NRUNS) / NRUNS
results[name].append(time)
ax = plt.axes()
lines = [ax.plot(X, Y, label=label)[0] for label, Y in results.items()]
ax.legend(handles=lines)
ax.set_xlabel("Percentage of values to take")
ax.set_title(f"Index length is {N}")
plt.savefig(f"nelem_index{N}.png") |
||
| # Relatively fast check allows us to not trigger `self.qc.get_axis()` computation | ||
| # if there're no negative indices and so they're not depend on the axis length. | ||
dchigarev marked this conversation as resolved.
Show resolved
Hide resolved
|
||
| if isinstance(axis_loc, np.ndarray) and not (axis_loc < 0).any(): | ||
| axis_lookup = axis_loc | ||
| else: | ||
| axis_lookup = pandas.RangeIndex(len(self.qc.get_axis(axis)))[ | ||
| axis_loc | ||
| ] | ||
|
|
||
| if isinstance(axis_lookup, pandas.Index) and not is_range_like(axis_lookup): | ||
| axis_lookup = axis_lookup.values | ||
| lookups.append(axis_lookup) | ||
| return lookups | ||
|
|
||
|
|
@@ -871,8 +1004,8 @@ def _check_dtypes(self, locator): | |
| """ | ||
| is_int = is_integer(locator) | ||
| is_int_slice = is_integer_slice(locator) | ||
| is_int_list = is_list_like(locator) and all(map(is_integer, locator)) | ||
| is_int_arr = is_integer_array(locator) | ||
| is_bool_arr = is_boolean_array(locator) | ||
|
|
||
| if not any([is_int, is_int_slice, is_int_list, is_bool_arr]): | ||
| if not any([is_int, is_int_slice, is_int_arr, is_bool_arr]): | ||
| raise ValueError(_ILOC_INT_ONLY_ERROR) | ||


There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Although
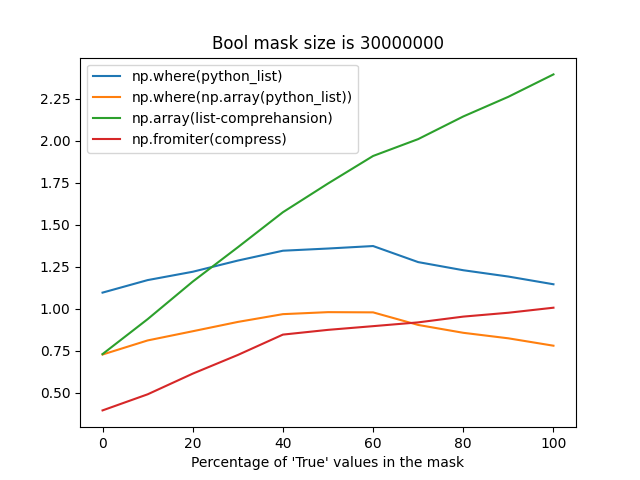
np.fromiterprobably suffers from reallocations when building an array from an iterator with an unknown size, it's still faster most of the time (when the percentage ofTruevalues in a mask is less than 75%).The difference between these strategies may take up to ~1 second for frames with the number of rows greater than 30 million, that's why I'm so picky about this place.
Time measurements
Bool mask size is 1_000_000:
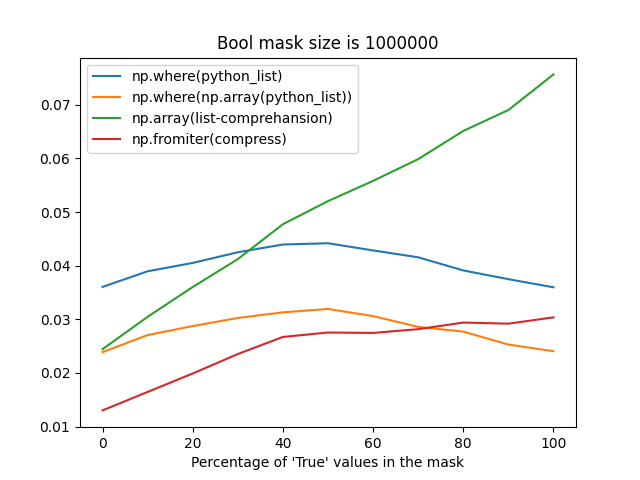
Bool mask size is 30_000_000:
Reproducer