-
Notifications
You must be signed in to change notification settings - Fork 653
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
FIX-#3686: optimize __getitem__ flow of loc/iloc
#3694
Conversation
Signed-off-by: Dmitry Chigarev <dmitry.chigarev@intel.com>
Codecov Report
@@ Coverage Diff @@
## master #3694 +/- ##
===========================================
- Coverage 85.47% 49.98% -35.50%
===========================================
Files 197 183 -14
Lines 16341 15690 -651
===========================================
- Hits 13968 7843 -6125
- Misses 2373 7847 +5474
Continue to review full report at Codecov.
|
…rray Signed-off-by: Dmitry Chigarev <dmitry.chigarev@intel.com>
| if any(i == -1 for i in row_lookup) or any(i == -1 for i in col_lookup): | ||
| raise KeyError( | ||
| "Passing list-likes to .loc or [] with any missing labels is no longer " | ||
| "supported, see https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#deprecate-loc-reindex-listlike" | ||
| ) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
this check was moved to the _compute_lookup
| if hasattr(result, "index") and isinstance(result.index, pandas.MultiIndex): | ||
| if ( | ||
| isinstance(result, (Series, DataFrame)) | ||
| and result._query_compiler.has_multiindex() |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
checking for MultiIndex with the special method helps to avoid delayed computations triggering in the case of OmniSci backend
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
this would be good as a code comment, not as a PR comment
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I'm not sure about this certain case. IMO this comment is excessive and would just bloat the if-statement. Any is_multiindex check has to be done with this special method, there's no something specific about its usage here that would make us add a comment.
How this if-statement looks with the comment added
# Option 1:
if (
isinstance(result, (Series, DataFrame))
# Checking for MultiIndex with the special method helps
# to avoid delayed computations triggering
and result._query_compiler.has_multiindex()
):
# Option 2:
if (
# Checking for MultiIndex with the special method helps
# to avoid delayed computations triggering
isinstance(result, (Series, DataFrame))
and result._query_compiler.has_multiindex()
):
# Option 3:
# Checking for MultiIndex with the special method helps
# to avoid delayed computations triggering
if (
isinstance(result, (Series, DataFrame))
and result._query_compiler.has_multiindex()
):| # It's faster to build the resulting numpy array from the reduced amount of data via | ||
| # `compress` iterator than convert non-numpy-like `indexer` to numpy and apply `np.where`. |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
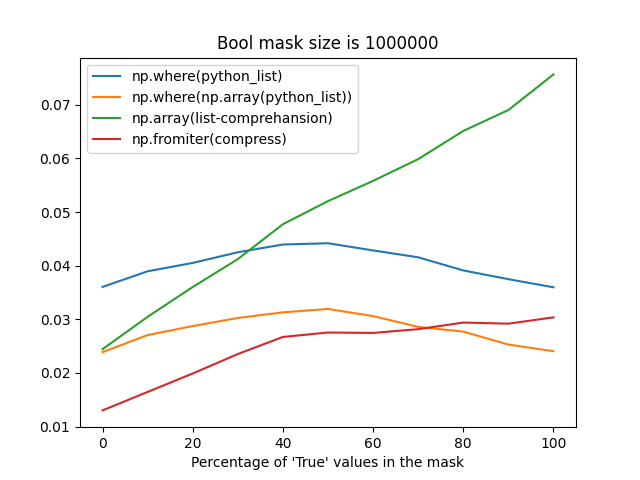
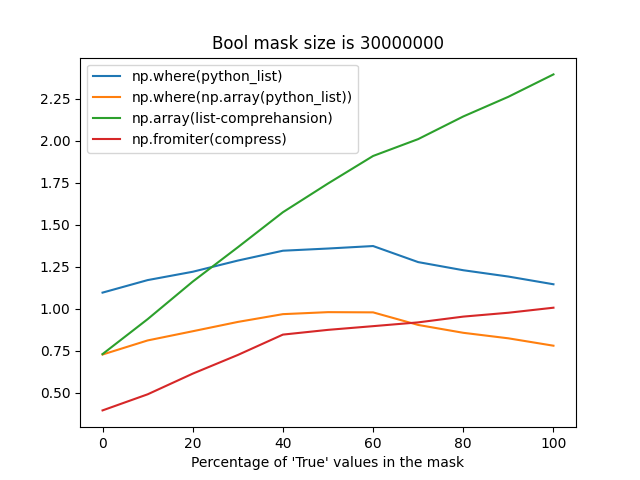
Although np.fromiter probably suffers from reallocations when building an array from an iterator with an unknown size, it's still faster most of the time (when the percentage of True values in a mask is less than 75%).
The difference between these strategies may take up to ~1 second for frames with the number of rows greater than 30 million, that's why I'm so picky about this place.
Reproducer
import numpy as np
import timeit
from itertools import compress
from matplotlib import pyplot as plt
N = 1_000_000
NRUNS = 5
def get_bool_python_list(n_elements, percentage_of_true_values):
number_of_true_vals = int(n_elements * percentage_of_true_values)
number_of_false_vals = int(n_elements * (1 - percentage_of_true_values))
bool_list = ([True] * number_of_true_vals) + ([False] * number_of_false_vals)
np.random.shuffle(bool_list)
return bool_list
fns = {
"np.where(python_list)": lambda arr: np.where(arr),
"np.where(np.array(python_list))": lambda arr: np.where(np.array(arr, dtype=bool)),
"np.array(list-comprehansion)": lambda arr: np.array(
[index for index, value in enumerate(arr) if value], dtype=np.int64,
),
"np.fromiter(compress)": lambda arr: np.fromiter(
compress(data=range(len(arr)), selectors=arr), dtype=np.int64
),
}
X = np.arange(0, 101, 10)
results = {key: [] for key in fns.keys()}
for x in X:
print(f"{x}%")
python_list = get_bool_python_list(n_elements=N, percentage_of_true_values=x / 100)
print(f"\tDEBUG: list length is {len(python_list)}")
for name, fn in fns.items():
print(f"\tRunning '{name}' fn...")
time = timeit.timeit(lambda: fn(python_list), number=NRUNS) / NRUNS
results[name].append(time)
ax = plt.axes()
lines = [ax.plot(X, Y, label=label)[0] for label, Y in results.items()]
ax.legend(handles=lines)
ax.set_xlabel("Percentage of 'True' values in the mask")
ax.set_title(f"Bool mask size is {N}")
plt.savefig(f"nelem{N}.png")| missing_labels = ( | ||
| # Converting `axis_loc` to maskable `np.array` to not fail | ||
| # on masking non-maskable list-like | ||
| np.array(axis_loc)[missing_mask] |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
axis_loc is always a numpy array in the current state of the above code, so this conversion is redundant. This is more about safety: if the code above would change, this error-reporting branch won't break anyway.
Signed-off-by: Dmitry Chigarev <dmitry.chigarev@intel.com>
92d082c
to
afd87af
Compare
| # Disabled for `BaseOnPython` because of the issue with `getitem_array`: | ||
| # https://github.com/modin-project/modin/issues/3701 | ||
| if get_current_execution() != "BaseOnPython": |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
.loc[series_boolean_mask] is now processed through the QueryCompiler.getitem_array method, and so affected by the same bug as the df[series_boolean_mask]
Signed-off-by: Dmitry Chigarev <dmitry.chigarev@intel.com>
Signed-off-by: Dmitry Chigarev <dmitry.chigarev@intel.com>
Signed-off-by: Dmitry Chigarev <dmitry.chigarev@intel.com>
a42f0e9
to
40885a3
Compare
Signed-off-by: Dmitry Chigarev <dmitry.chigarev@intel.com>
bf3384f
to
bf042ca
Compare
| # Disabled for `BaseOnPython` because of the issue with `getitem_array`. | ||
| # groupby.shift internally masks the source frame with a Series boolean mask, | ||
| # doing so ends up in the `getitem_array` method, that is broken for `BaseOnPython`: | ||
| # https://github.com/modin-project/modin/issues/3701 |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Boolean masking inside shift is done here
| # `Index.__getitem__` works much faster with numpy arrays than with python lists, | ||
| # so although we lose some time here on converting to numpy, `Index.__getitem__` | ||
| # speedup covers the loss that we gain here. | ||
| axis_loc = np.array(axis_loc, dtype=np.int64) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Reproducer
import numpy as np
import timeit
from matplotlib import pyplot as plt
import pandas
N = 1_000_000
NRUNS = 5
index = pandas.RangeIndex(N)
def get_positions(index, percentage):
n_elements = int(len(index) * percentage)
indexer = list(range(n_elements))
return list(np.random.choice(indexer, n_elements, replace=False))
fns = {
"RangeIndex.__getitem__(python_list)": lambda index, indexer: index[indexer],
"RangeIndex.__getitem__(np.array(python_list))": lambda index, indexer: index[
np.array(indexer, dtype=np.int64)
],
}
X = np.arange(0, 101, 10)
results = {key: [] for key in fns.keys()}
for x in X:
print(f"{x}%")
python_list = get_positions(index=index, percentage=x / 100)
print(f"\tDEBUG: list length is {len(python_list)}")
for name, fn in fns.items():
print(f"\tRunning '{name}' fn...")
time = timeit.timeit(lambda: fn(index, python_list), number=NRUNS) / NRUNS
results[name].append(time)
ax = plt.axes()
lines = [ax.plot(X, Y, label=label)[0] for label, Y in results.items()]
ax.legend(handles=lines)
ax.set_xlabel("Percentage of values to take")
ax.set_title(f"Index length is {N}")
plt.savefig(f"nelem_index{N}.png")| if hasattr(result, "index") and isinstance(result.index, pandas.MultiIndex): | ||
| if ( | ||
| isinstance(result, (Series, DataFrame)) | ||
| and result._query_compiler.has_multiindex() |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
this would be good as a code comment, not as a PR comment
Signed-off-by: Dmitry Chigarev <dmitry.chigarev@intel.com>
Signed-off-by: Dmitry Chigarev <dmitry.chigarev@intel.com>
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
LGTM, will put green check when tests pass.
| if isinstance(row_loc, slice) and row_loc == slice(None): | ||
| # If we're only slicing columns, handle the case with `__getitem__` | ||
| if not isinstance(col_loc, slice): | ||
| # Boolean indexers can just be sliced into the columns object and | ||
| # then passed to `__getitem__` | ||
| if is_boolean_array(col_loc): | ||
| col_loc = self.df.columns[col_loc] | ||
| return self.df.__getitem__(col_loc) | ||
| else: | ||
| result_slice = self.df.columns.slice_locs(col_loc.start, col_loc.stop) | ||
| return self.df.iloc[:, slice(*result_slice)] |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
This is no longer needed, now the natural flow of .loc/.iloc performs the same as this fast path.
This comment is no longer relevant:
Shape: (5000, 5000)
__getitem__: 0.7248090452048928
.loc: 0.6661503429058939
.loc: 0.6549520299304277
__getitem__: 0.6364482759963721
Shape: (10000000, 10)
__getitem__: 0.6415216489695013
.loc: 0.6545198771636933
.loc: 0.6536120360251516
__getitem__: 0.6337228799238801
Reproducer
import modin.pandas as pd
import numpy as np
import timeit
shapes = [
# Wide frame
(5000, 5000),
# Long frame
(10_000_000, 10),
]
NRUNS = 100
for shape in shapes:
NROWS, NCOLS = shape
df = pd.DataFrame({f"col{i}": np.arange(NROWS) for i in range(NCOLS)})
print(f"Shape: {shape}")
print(f"__getitem__: {timeit.timeit(lambda: repr(df[['col1']]), number=NRUNS)}")
print(f".loc: {timeit.timeit(lambda: repr(df.loc[:, ['col1']]), number=NRUNS)}")
print(f".loc: {timeit.timeit(lambda: repr(df.loc[:, ['col1']]), number=NRUNS)}")
print(f"__getitem__: {timeit.timeit(lambda: repr(df[['col1']]), number=NRUNS)}")Also, check the results of ASV runs with the __getitem__ benchmarks added at the PR's header.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
LGTM, thanks @dchigarev!
|
We will merge this tomorrow (24h after this post) if no one else from @modin-project/modin-core has more comments. |



Signed-off-by: Dmitry Chigarev dmitry.chigarev@intel.com
What do these changes do?
flake8 modin/ asv_bench/benchmarks scripts/doc_checker.pyblack --check modin/ asv_bench/benchmarks scripts/doc_checker.pygit commit -s__getitem__flow ofloc/iloc#3686docs/developer/architecture.rstis up-to-dateResults of ASV benchmarks (existed ones were combined with the added in #3700):
PandasOnRay execution
Launch command:
Note: the regression in the case of "bool_python_list" is caused by the need of converting a python boolean mask to a numpy array (it wasn't done before).
OmnisciOnNative execution
Launch command: