-
Notifications
You must be signed in to change notification settings - Fork 2
Custom Analysis Part 7
And finally, I can have a look at my cells. Does it make sense?
First, I’ll visualize the data we used for the clustering: the mean
fluorescence over time. I use ggplot2 for this.
library(ggplot2)
ggplot(cells_mean) +
geom_path(aes(x=percentage,
y=x,
group=celldiv)) +
facet_grid(~COR)
As you see, it’s a bit messy, but it seems like in the first group, fluorescence goes up, in the second it goes down, and in the last it stays more or less the same. I’ll make the plot a bit easier to look at (note that I use the same color as I used in the DnaX/FtsZ trajectory plots of the previous tutorial):
reddish <- "#B63679FF"
ggplot(cells_mean) +
geom_path(aes(x=percentage,
y=x,
group=celldiv),
alpha=0.4) +
facet_grid(~COR) +
theme_minimal() +
geom_smooth(aes(x=percentage,
y=x),
color=reddish,
fill=reddish) +
ylab("Mean fluorescence intensity (AU)")
Now, let’s have a look at the kymographs of the three groups by division percentage.
#make a list of three kymo plots based on COR
threeKymos <- lapply(c(1:3), function(y)
bactKymo(cells_image_grouped[cells_image_grouped$COR==y,], percDiv=TRUE, sizeAV=TRUE, timeD=TRUE)
)
#use arrangeGrob to plot them all on one page
plot(gridExtra::arrangeGrob(grobs=threeKymos, nrow=1))
Now, finally, let’s have a look at the single kymographs of each group. For this, I put them all in one big PDF.
listofgroupedkymos <- lapply(c(1:3), function(y)
gridExtra::arrangeGrob(
grobs=bactKymo(cells_image_grouped[cells_image_grouped$COR==y,], sizeAV=TRUE, timeD=TRUE, legend=FALSE)
)
)
cairo_pdf(filename="kymo-per-group.pdf", onefile=TRUE, width=20, height=20, pointsize=4)
lapply(listofgroupedkymos, function(x) plot(x))
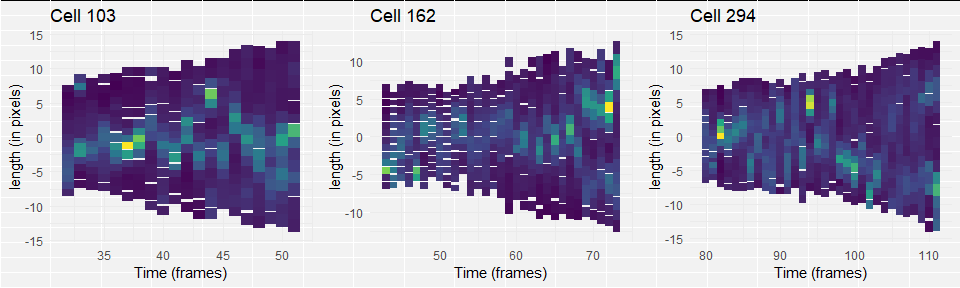
dev.off()When looking at the groups, it seems like in group one, the replisome dissociates towards the end of the cell cycle and re-appears on one or two sides of the cell. In some instances, however, there is no clear association. In group 2, there is no or low fluorescence at the beginning of the cell, but after that two replisomes associate and stay until the end of the cycle. I think that in this group, association and dissociation happens to quickly after each other to see within the 2 minute interval. In the last group, no clear dissociation, splitting, or association takes place. These cells could be sick, but another possibility is that the moment of “real” cell division is determined wrongly by the segmentation program. This could be tested by imaging the replisome together with a membrane marker.
I finally pick three group members for the plots of my figure. Group 1: cell 294, group 2: cell 162, group 3: cell 103.
plot(gridExtra::arrangeGrob(
grobs = bactKymo(cells_image_grouped[cells_image_grouped$cell%in%c(294, 162, 103),],
sizeAV=TRUE,
timeD=TRUE,
bins=16,
legend=FALSE),
nrow=1)
)
| ⬅️ Custom Analysis Part 6: Cluster Analysis | Custom Analysis Part 8: Resources ➡️ |
|---|
