-
Notifications
You must be signed in to change notification settings - Fork 4
FEAT2FS
This tutorial is fantastic https://surfer.nmr.mgh.harvard.edu/fswiki/FsTutorial/FslFeatFreeSurfer
Basically:
- Register the functionals to the freesurfer's T1 volume.
reg-feat2anat --feat /path/to/analysis.feat --subject subjectID
If registration to std space has not been performed, you need to do it first. If it is a gfeat, then you may need to copy the ''/reg'' directory from the first level analysis.
However, step 1 was failing for me, specifically during reg-feat2anat. I tracked the bug to this: [[http://www.mail-archive.com/freesurfer@nmr.mgh.harvard.edu/msg29122.html]]
All that is needed to fix the registration bug is to replace the script as provided in the above link. The only difference between the old version and the new one is that the new one specifies the file suffix, while the old one does not. Stupid bug, I lost a whole morning on this.
While you can see the volume overlaid on the surface with tksurfer, as the tutorial suggests, freeview works much smoother.
First, load the backround, subject-space t1 and surface
freeview -v subject/mri/brain.mgz -f subject/surf/lh.white
now, load the overlay, but taking care to specify the registration needed to go from fMRI subject space to T1 subject space. The registratin is featDirectory ''/reg/freesurfer/anat2exf.register.dat''
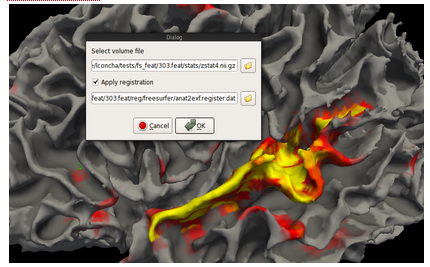
And, to load data that is in standard space onto the subjects surface, for example after comparing two copes of the same subject using a fixed effects model, we need another transformation, this time anat2std.register.dat.
That same transformation can also be used to load the volumes (e.g. zstat1.nii.gz) to overlay on top of brain.mgz of the subject.
Tabla de contenidos
- Home
- Como colaborar en la Wiki
- rocket.chat
- Resonadores
- Bash
- Clúster
- Procesamiento de Imágenes
- fMRI
- DW-MRI
- FIJI - Análisis histológico
- Herramientas Software
- Otros